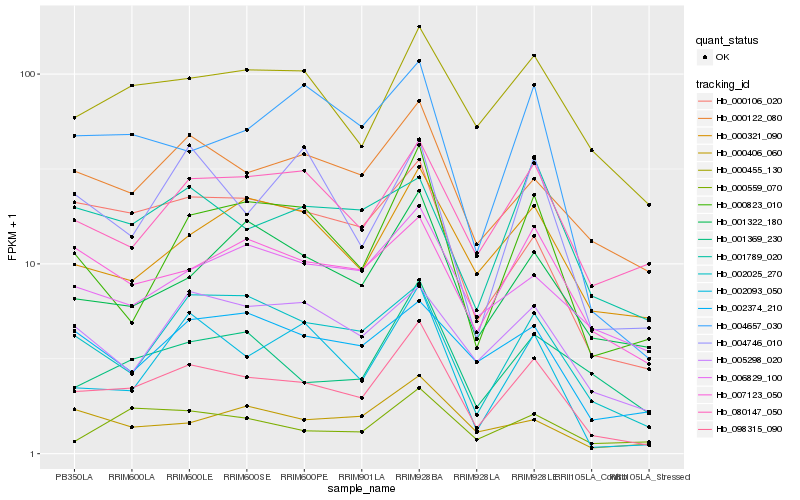
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_098315_090 |
0.0 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 2 |
Hb_000823_010 |
0.1336586079 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 3 |
Hb_000106_020 |
0.1341524975 |
- |
- |
PREDICTED: protein TOPLESS isoform X1 [Jatropha curcas] |
| 4 |
Hb_000122_080 |
0.1374425046 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 5 |
Hb_002025_270 |
0.1381467717 |
- |
- |
PREDICTED: uncharacterized protein At4g15970 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004657_030 |
0.140851559 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 7 |
Hb_000455_130 |
0.1421061598 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002093_050 |
0.1436791236 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 9 |
Hb_007123_050 |
0.1480193786 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 10 |
Hb_001322_180 |
0.1507191432 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 11 |
Hb_001369_230 |
0.1511082185 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 12 |
Hb_080147_050 |
0.1514855762 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 13 |
Hb_000321_090 |
0.1538371553 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 14 |
Hb_000559_070 |
0.1539976445 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 15 |
Hb_004746_010 |
0.1546164947 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 16 |
Hb_001789_020 |
0.1558890248 |
- |
- |
PREDICTED: tripeptidyl-peptidase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000406_060 |
0.1573512763 |
- |
- |
hypothetical protein CICLE_v10000115mg [Citrus clementina] |
| 18 |
Hb_005298_020 |
0.157945553 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 2 [Jatropha curcas] |
| 19 |
Hb_006829_100 |
0.160061553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002374_210 |
0.1610215082 |
- |
- |
PREDICTED: uncharacterized protein LOC105638976 [Jatropha curcas] |