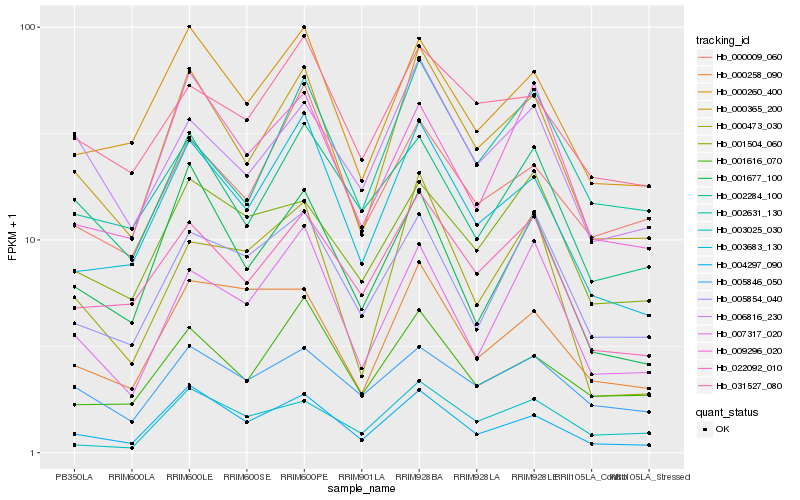
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_200 |
0.0 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 2 |
Hb_003683_130 |
0.0865931344 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004297_090 |
0.0906940245 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 4 |
Hb_000260_400 |
0.0999619649 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 5 |
Hb_022092_010 |
0.100245041 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 6 |
Hb_001677_100 |
0.1145047241 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 7 |
Hb_006816_230 |
0.119459818 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 8 |
Hb_001616_070 |
0.1198129042 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 9 |
Hb_002284_100 |
0.1236157157 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 10 |
Hb_007317_020 |
0.1243714686 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009296_020 |
0.127325093 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_002631_130 |
0.1289489042 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 13 |
Hb_031527_080 |
0.1293481388 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000473_030 |
0.1300638715 |
- |
- |
hypothetical protein JCGZ_19297 [Jatropha curcas] |
| 15 |
Hb_005846_050 |
0.130636201 |
- |
- |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 16 |
Hb_000258_090 |
0.1329981613 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 17 |
Hb_005854_040 |
0.1338303982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001504_060 |
0.1365437312 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 19 |
Hb_000009_060 |
0.137786978 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 20 |
Hb_003025_030 |
0.137793286 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |