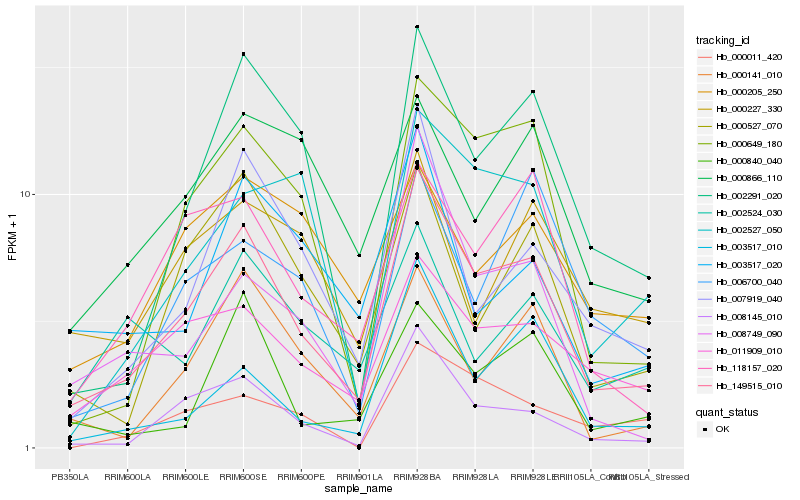
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_149515_010 |
0.0 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 2 |
Hb_008145_010 |
0.1457134953 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 3 |
Hb_011909_010 |
0.1551341327 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 4 |
Hb_000649_180 |
0.1568467677 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 5 |
Hb_002291_020 |
0.1579552009 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_007919_040 |
0.1653647163 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |
| 7 |
Hb_000141_010 |
0.1709047043 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 8 |
Hb_118157_020 |
0.1709709301 |
- |
- |
hypothetical protein MTR_3g456110 [Medicago truncatula] |
| 9 |
Hb_000527_070 |
0.1720481671 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 10 |
Hb_008749_090 |
0.1806250729 |
- |
- |
hypothetical protein CICLE_v10017161mg [Citrus clementina] |
| 11 |
Hb_003517_010 |
0.1865964081 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000840_040 |
0.1867577692 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 13 |
Hb_003517_020 |
0.189596002 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 14 |
Hb_002527_050 |
0.1941162268 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 15 |
Hb_000227_330 |
0.1968475954 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 16 |
Hb_000866_110 |
0.1977182133 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 17 |
Hb_006700_040 |
0.1989733687 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 18 |
Hb_002524_030 |
0.1991561481 |
- |
- |
- |
| 19 |
Hb_000011_420 |
0.1993313992 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g29660-like [Citrus sinensis] |
| 20 |
Hb_000205_250 |
0.200944827 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |