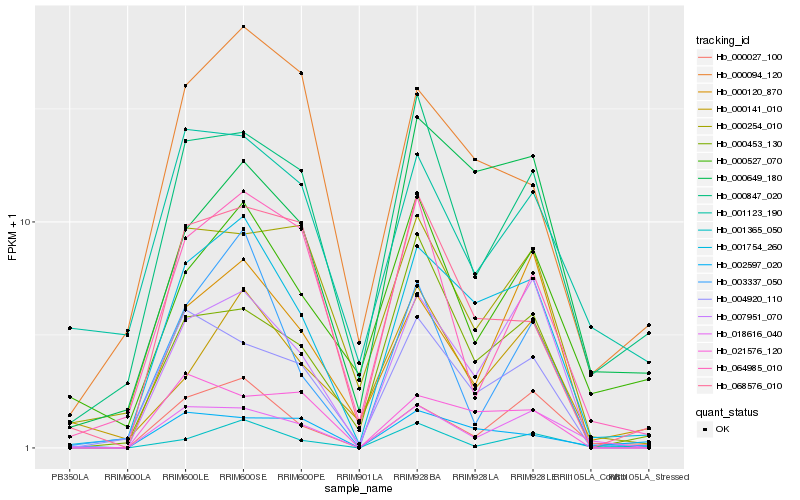
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001754_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643409 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007951_070 |
0.161347434 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 3 |
Hb_000027_100 |
0.1702006329 |
- |
- |
hypothetical protein VITISV_039936 [Vitis vinifera] |
| 4 |
Hb_004920_110 |
0.1802126552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000847_020 |
0.1828634446 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 6 |
Hb_018616_040 |
0.1839524121 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000094_120 |
0.1868210009 |
- |
- |
PREDICTED: uncharacterized protein At1g01500 [Jatropha curcas] |
| 8 |
Hb_000649_180 |
0.1892472764 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 9 |
Hb_003337_050 |
0.1999831215 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 10 |
Hb_000527_070 |
0.2032202788 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 11 |
Hb_000120_870 |
0.2033635639 |
rubber biosynthesis |
Gene Name: CPT6 |
- |
| 12 |
Hb_000254_010 |
0.2052322464 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 13 |
Hb_068576_010 |
0.2062513196 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 14 |
Hb_021576_120 |
0.2066673183 |
- |
- |
DNA-directed RNA polymerase subunit, putative [Ricinus communis] |
| 15 |
Hb_000453_130 |
0.2093407933 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_000141_010 |
0.2101841638 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 17 |
Hb_001123_190 |
0.2122395662 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 18 |
Hb_064985_010 |
0.2125810101 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 19 |
Hb_002597_020 |
0.2155901274 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 20 |
Hb_001365_050 |
0.2165296458 |
- |
- |
hypothetical protein POPTR_0019s128601g, partial [Populus trichocarpa] |