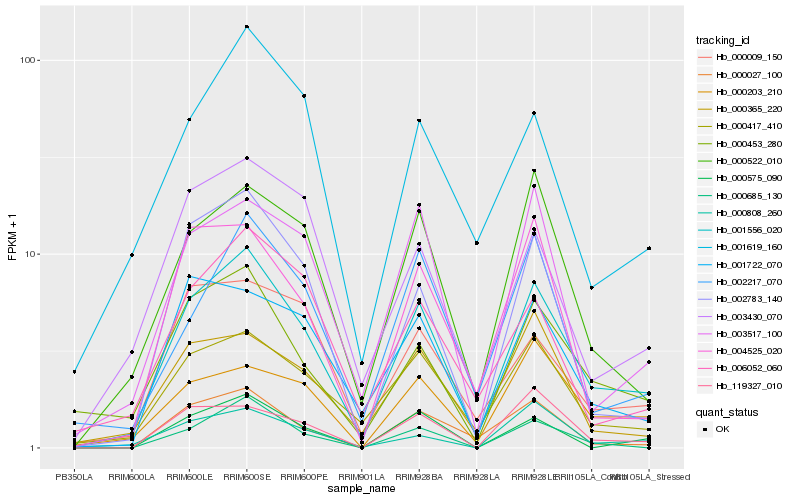
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001556_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 2 |
Hb_000417_410 |
0.1349656196 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000453_280 |
0.1445178078 |
- |
- |
negative cofactor 2 transcriptional co-repressor, putative [Ricinus communis] |
| 4 |
Hb_000522_010 |
0.1450476991 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 5 |
Hb_002783_140 |
0.1490654553 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 6 |
Hb_003517_100 |
0.1552426489 |
transcription factor |
TF Family: GRAS |
- |
| 7 |
Hb_000027_100 |
0.1578198777 |
- |
- |
hypothetical protein VITISV_039936 [Vitis vinifera] |
| 8 |
Hb_002217_070 |
0.1639646355 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 9 |
Hb_001722_070 |
0.1642604377 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 10 |
Hb_000365_220 |
0.1642628943 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 11 |
Hb_004525_020 |
0.1679374474 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 12 |
Hb_001619_160 |
0.1688363532 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 13 |
Hb_000203_210 |
0.1691193233 |
- |
- |
PREDICTED: uncharacterized protein LOC105040619 isoform X3 [Elaeis guineensis] |
| 14 |
Hb_119327_010 |
0.1703752614 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 15 |
Hb_000685_130 |
0.1706452088 |
- |
- |
PREDICTED: uncharacterized protein LOC105351820 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_006052_060 |
0.1709746856 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 17 |
Hb_000575_090 |
0.1716045308 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 24 [Populus euphratica] |
| 18 |
Hb_000808_260 |
0.1727291305 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 19 |
Hb_000009_150 |
0.1730499936 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 20 |
Hb_003430_070 |
0.1733393726 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |