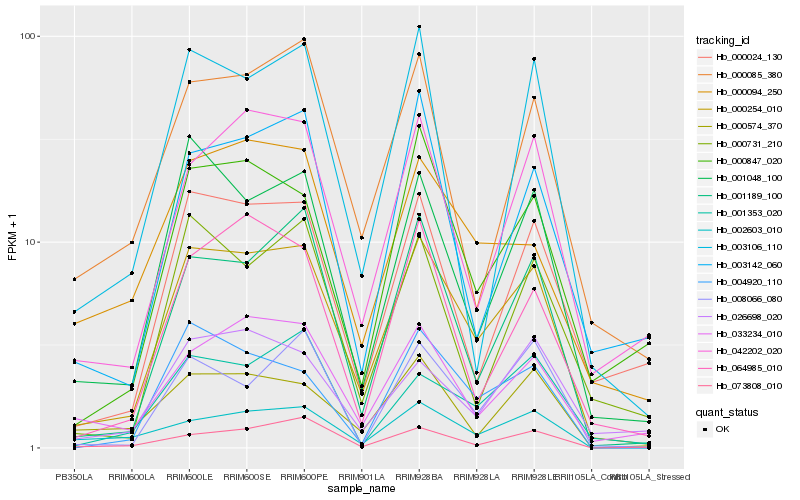
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000254_010 |
0.0 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 2 |
Hb_001048_100 |
0.1299987096 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 3 |
Hb_001189_100 |
0.130111422 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Jatropha curcas] |
| 4 |
Hb_042202_020 |
0.1391601992 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 5 |
Hb_033234_010 |
0.1403921304 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 6 |
Hb_000847_020 |
0.143688323 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 7 |
Hb_000024_130 |
0.1445402755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000085_380 |
0.1458892778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004920_110 |
0.1489384047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002603_010 |
0.1509545146 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_003106_110 |
0.1513192163 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_064985_010 |
0.1554144495 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 13 |
Hb_073808_010 |
0.1560636824 |
desease resistance |
Gene Name: DUF594 |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000574_370 |
0.1561415725 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 15 |
Hb_001353_020 |
0.1573758828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_026698_020 |
0.1578190151 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 17 |
Hb_008066_080 |
0.1591516396 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 18 |
Hb_000731_210 |
0.1592917012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000094_250 |
0.1613378909 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X3 [Jatropha curcas] |
| 20 |
Hb_003142_060 |
0.1615320078 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |