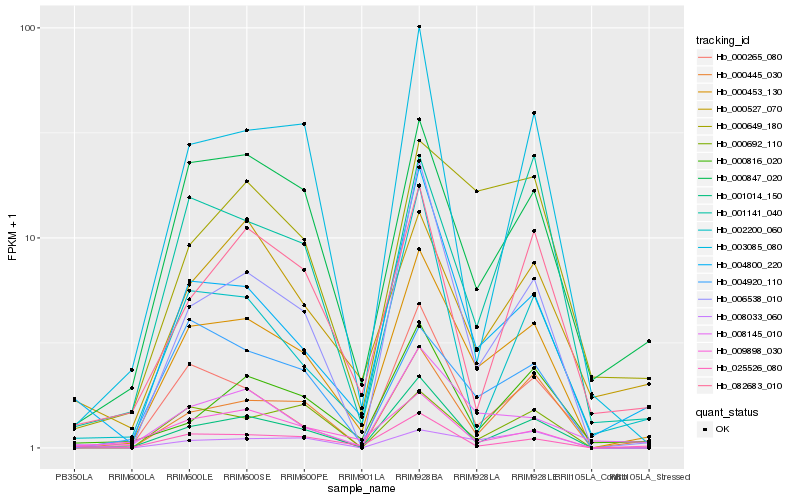
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_130 |
0.0 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_001014_150 |
0.1501026826 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000847_020 |
0.15236244 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 4 |
Hb_006538_010 |
0.1593944127 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |
| 5 |
Hb_004800_220 |
0.1655872541 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 6 |
Hb_008145_010 |
0.169118754 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 7 |
Hb_000445_030 |
0.1697461319 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 8 |
Hb_025526_080 |
0.1706332018 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Vitis vinifera] |
| 9 |
Hb_000265_080 |
0.1783940895 |
- |
- |
Vacuolar cation/proton exchanger, putative [Ricinus communis] |
| 10 |
Hb_004920_110 |
0.1844562978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001141_040 |
0.1846973079 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 12 |
Hb_000649_180 |
0.1853259948 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 13 |
Hb_003085_080 |
0.1900488101 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_008033_060 |
0.1909157091 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 15 |
Hb_000816_020 |
0.197225792 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_009898_030 |
0.1990002969 |
- |
- |
PREDICTED: uncharacterized protein LOC103720281 [Phoenix dactylifera] |
| 17 |
Hb_000692_110 |
0.201320377 |
- |
- |
PREDICTED: NEP1-interacting protein-like 1 [Jatropha curcas] |
| 18 |
Hb_000527_070 |
0.2023312597 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 19 |
Hb_082683_010 |
0.2029625145 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_002200_060 |
0.2058267039 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |