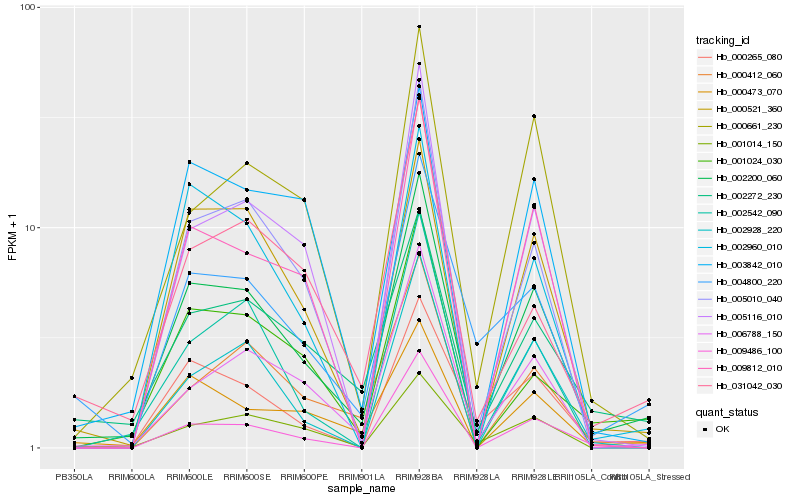
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002200_060 |
0.0 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 2 |
Hb_000521_360 |
0.1325152718 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 3 |
Hb_000473_070 |
0.1335498406 |
- |
- |
PREDICTED: amino-acid permease BAT1 homolog [Jatropha curcas] |
| 4 |
Hb_002272_230 |
0.1369301048 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 5 |
Hb_000265_080 |
0.1384586643 |
- |
- |
Vacuolar cation/proton exchanger, putative [Ricinus communis] |
| 6 |
Hb_006788_150 |
0.1385797892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004800_220 |
0.1445577717 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 8 |
Hb_005010_040 |
0.1446616214 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 9 |
Hb_001024_030 |
0.1467359917 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_031042_030 |
0.1516791449 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_009812_010 |
0.1529040664 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_001014_150 |
0.1535363992 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_000661_230 |
0.1550405262 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000412_060 |
0.1554590822 |
- |
- |
hypothetical protein JCGZ_18814 [Jatropha curcas] |
| 15 |
Hb_005116_010 |
0.1585982158 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 16 |
Hb_002542_090 |
0.1591907794 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 17 |
Hb_002960_010 |
0.1593672633 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 18 |
Hb_002928_220 |
0.1606613994 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 19 |
Hb_009486_100 |
0.1609753936 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_003842_010 |
0.1617016111 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |