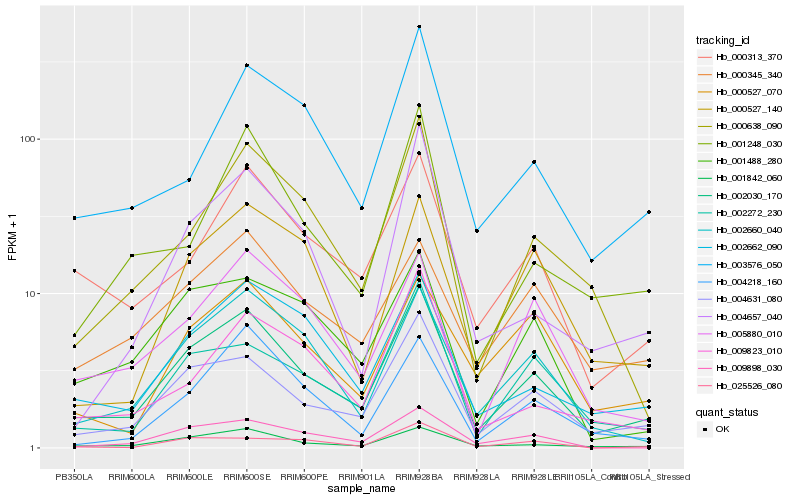
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002030_170 |
0.0 |
- |
- |
PREDICTED: calcium-transporting ATPase 1-like [Jatropha curcas] |
| 2 |
Hb_004631_080 |
0.0900590896 |
- |
- |
P3B-ATPase PH1 [Petunia x hybrida] |
| 3 |
Hb_001842_060 |
0.1056908511 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 4 |
Hb_002660_040 |
0.1361884104 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002272_230 |
0.1381376801 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 6 |
Hb_000313_370 |
0.1382581926 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_002662_090 |
0.1392162166 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 8 |
Hb_001488_280 |
0.1528216237 |
- |
- |
hypothetical protein JCGZ_19947 [Jatropha curcas] |
| 9 |
Hb_005880_010 |
0.1542024135 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 10 |
Hb_000638_090 |
0.1542153744 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_009898_030 |
0.1550092462 |
- |
- |
PREDICTED: uncharacterized protein LOC103720281 [Phoenix dactylifera] |
| 12 |
Hb_003576_050 |
0.1592554296 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 13 |
Hb_000345_340 |
0.1603054479 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 14 |
Hb_004218_160 |
0.1614565599 |
- |
- |
NBS-LRR type disease resistance protein [Medicago truncatula] |
| 15 |
Hb_001248_030 |
0.163275922 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000527_070 |
0.1654596609 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 17 |
Hb_025526_080 |
0.1656021851 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Vitis vinifera] |
| 18 |
Hb_000527_140 |
0.1663469933 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 19 |
Hb_009823_010 |
0.1695677476 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 20 |
Hb_004657_040 |
0.1712062575 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |