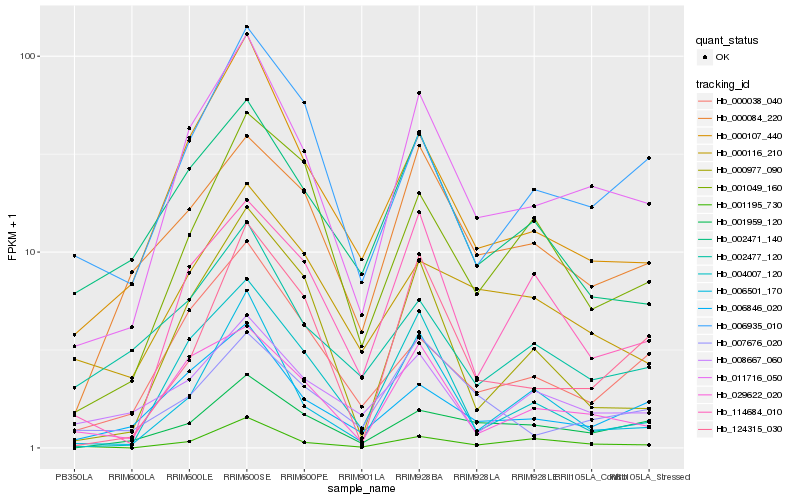
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011716_050 |
0.0 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 2 |
Hb_000107_440 |
0.1307873248 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 3 |
Hb_029622_020 |
0.1389018386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006846_020 |
0.1394580366 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 5 |
Hb_000038_040 |
0.1510857246 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 6 |
Hb_006935_010 |
0.1591743269 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 7 |
Hb_001959_120 |
0.1625954056 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 8 |
Hb_000084_220 |
0.1667011827 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 9 |
Hb_001195_730 |
0.1670646447 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 10 |
Hb_114684_010 |
0.1706570478 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 11 |
Hb_000116_210 |
0.1761356385 |
- |
- |
PREDICTED: uncharacterized protein LOC105628567 [Jatropha curcas] |
| 12 |
Hb_002471_140 |
0.1761589355 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 13 |
Hb_008667_060 |
0.1769417357 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 14 |
Hb_004007_120 |
0.177231507 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type isoform X1 [Populus euphratica] |
| 15 |
Hb_001049_160 |
0.1786987947 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 16 |
Hb_124315_030 |
0.1793176766 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 17 |
Hb_000977_090 |
0.1816900926 |
- |
- |
hypothetical protein POPTR_0010s18550g [Populus trichocarpa] |
| 18 |
Hb_002477_120 |
0.1824369073 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 19 |
Hb_007676_020 |
0.1849102397 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006501_170 |
0.1862596401 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |