| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
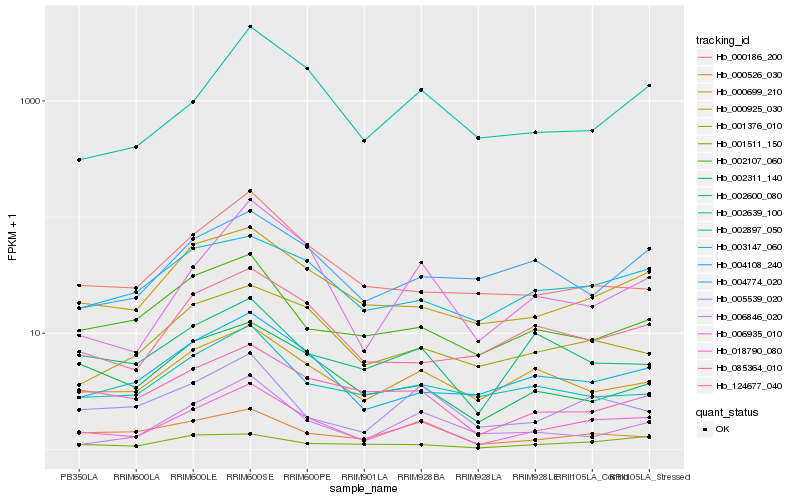
Hb_018790_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s06335g [Populus trichocarpa] |
| 2 |
Hb_000925_030 |
0.1455158763 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 3 |
Hb_004108_240 |
0.1455272545 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 4 |
Hb_000699_210 |
0.1529685411 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 5 |
Hb_006935_010 |
0.1546490718 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 6 |
Hb_005539_020 |
0.1579628382 |
- |
- |
- |
| 7 |
Hb_003147_060 |
0.1589978497 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 8 |
Hb_124677_040 |
0.1612506337 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 9 |
Hb_002639_100 |
0.1638317336 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 10 |
Hb_001376_010 |
0.167619272 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 11 |
Hb_001511_150 |
0.1724014401 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 12 |
Hb_002897_050 |
0.1732818362 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 13 |
Hb_002600_080 |
0.1742517896 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_002107_060 |
0.1745750283 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 15 |
Hb_000526_030 |
0.1751204999 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 16 |
Hb_000186_200 |
0.1775500122 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 17 |
Hb_002311_140 |
0.1784314256 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 18 |
Hb_085364_010 |
0.178830471 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 19 |
Hb_006846_020 |
0.1799071215 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 20 |
Hb_004774_020 |
0.1801238807 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |