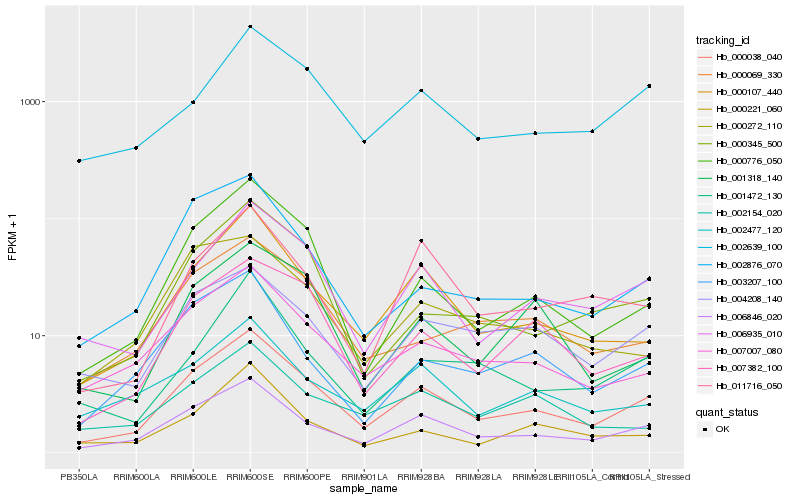
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000038_040 |
0.0 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 2 |
Hb_006846_020 |
0.0731632061 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 3 |
Hb_006935_010 |
0.1092299119 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 4 |
Hb_000107_440 |
0.1201489996 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 5 |
Hb_007007_080 |
0.1240217275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000776_050 |
0.1338201099 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 7 |
Hb_007382_100 |
0.1355813636 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_004208_140 |
0.1382948519 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 9 |
Hb_000345_500 |
0.1384249166 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 10 |
Hb_002639_100 |
0.1408837665 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 11 |
Hb_000069_330 |
0.1462618407 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_002477_120 |
0.147836437 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_002876_070 |
0.1500923875 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 14 |
Hb_011716_050 |
0.1510857246 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 15 |
Hb_003207_100 |
0.1533627823 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP9-like [Jatropha curcas] |
| 16 |
Hb_001472_130 |
0.1553030268 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 17 |
Hb_001318_140 |
0.1579498775 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 18 |
Hb_002154_020 |
0.1579830547 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_000221_060 |
0.1584193592 |
- |
- |
- |
| 20 |
Hb_000272_110 |
0.1615207532 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |