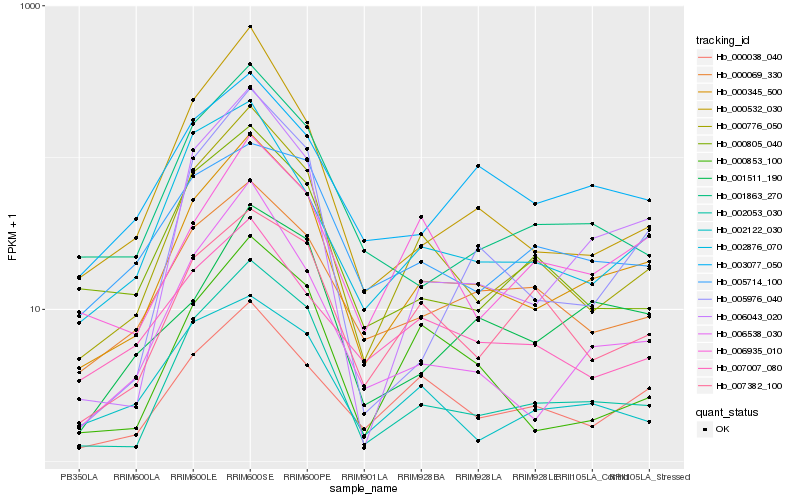
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_500 |
0.0 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 2 |
Hb_002053_030 |
0.108117652 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000776_050 |
0.1085363954 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 4 |
Hb_006043_020 |
0.1319122009 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |
| 5 |
Hb_006538_030 |
0.131948101 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 6 |
Hb_000069_330 |
0.1358935117 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_001863_270 |
0.1374042232 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 8 |
Hb_000038_040 |
0.1384249166 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 9 |
Hb_002876_070 |
0.1410940607 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 10 |
Hb_003077_050 |
0.1441298327 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 11 |
Hb_000532_030 |
0.1497079408 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |
| 12 |
Hb_001511_190 |
0.1513847949 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 13 |
Hb_000805_040 |
0.1540570073 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 14 |
Hb_006935_010 |
0.1615189968 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 15 |
Hb_007382_100 |
0.1626938073 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000853_100 |
0.1636122934 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |
| 17 |
Hb_005976_040 |
0.1638052454 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 18 |
Hb_002122_030 |
0.1653115789 |
- |
- |
- |
| 19 |
Hb_007007_080 |
0.1658190798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005714_100 |
0.1675847382 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |