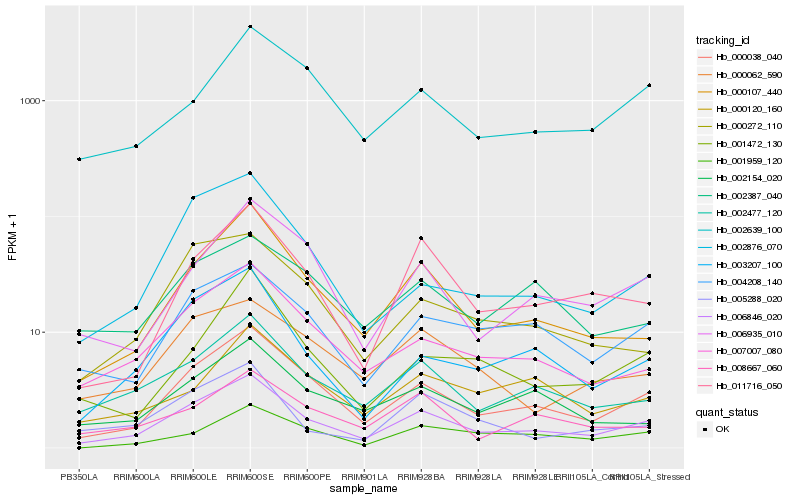
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006846_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 2 |
Hb_000038_040 |
0.0731632061 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 3 |
Hb_002477_120 |
0.1204342913 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_007007_080 |
0.1244369601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000107_440 |
0.1257835136 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 6 |
Hb_004208_140 |
0.1298466792 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 7 |
Hb_003207_100 |
0.1373936727 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP9-like [Jatropha curcas] |
| 8 |
Hb_011716_050 |
0.1394580366 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 9 |
Hb_006935_010 |
0.1437590617 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 10 |
Hb_000062_590 |
0.1530938193 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_002639_100 |
0.1556545234 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 12 |
Hb_002154_020 |
0.1559951131 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000272_110 |
0.1576485956 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002876_070 |
0.1620664758 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 15 |
Hb_005288_020 |
0.1647443569 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_008667_060 |
0.1660402675 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 17 |
Hb_001472_130 |
0.1671303443 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 18 |
Hb_000120_160 |
0.167209651 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_001959_120 |
0.1701391981 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 20 |
Hb_002387_040 |
0.1717240595 |
- |
- |
protein transporter, putative [Ricinus communis] |