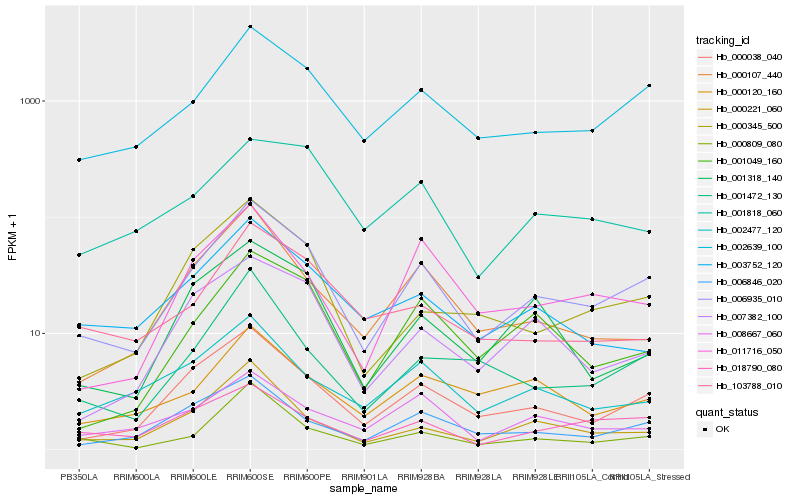
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006935_010 |
0.0 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 2 |
Hb_002639_100 |
0.1018764616 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 3 |
Hb_000038_040 |
0.1092299119 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 4 |
Hb_006846_020 |
0.1437590617 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 5 |
Hb_001049_160 |
0.1478926708 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 6 |
Hb_007382_100 |
0.1537348822 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 7 |
Hb_018790_080 |
0.1546490718 |
- |
- |
hypothetical protein POPTR_0011s06335g [Populus trichocarpa] |
| 8 |
Hb_000120_160 |
0.1584003744 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000221_060 |
0.1587004218 |
- |
- |
- |
| 10 |
Hb_011716_050 |
0.1591743269 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 11 |
Hb_000107_440 |
0.159804729 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 12 |
Hb_008667_060 |
0.1606237358 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 13 |
Hb_000809_080 |
0.1607651238 |
- |
- |
50S ribosomal protein L25, putative [Ricinus communis] |
| 14 |
Hb_000345_500 |
0.1615189968 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 15 |
Hb_001472_130 |
0.1623911076 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 16 |
Hb_002477_120 |
0.1632828213 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 17 |
Hb_001318_140 |
0.165354539 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 18 |
Hb_003752_120 |
0.1657560826 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001818_060 |
0.1662201166 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 20 |
Hb_103788_010 |
0.1664469992 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |