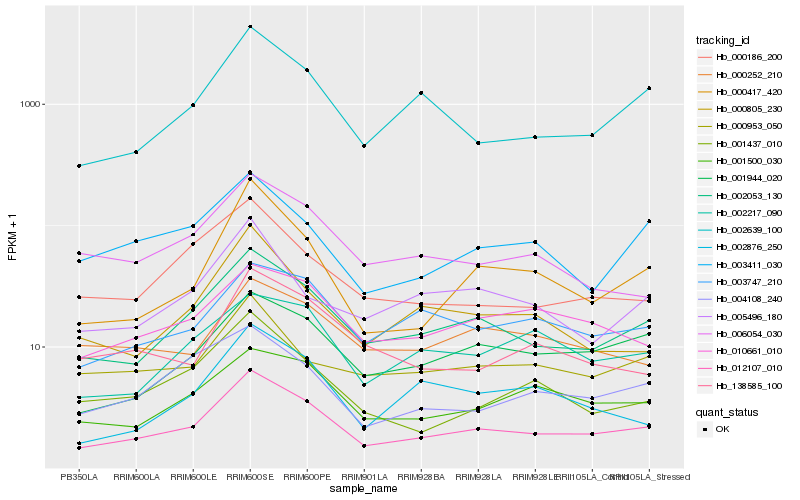
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012107_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 2 |
Hb_002053_130 |
0.0760417463 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 3 |
Hb_001944_020 |
0.1168696832 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 4 |
Hb_000417_420 |
0.1191908261 |
- |
- |
PREDICTED: U-box domain-containing protein 17 [Jatropha curcas] |
| 5 |
Hb_002639_100 |
0.1205606122 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 6 |
Hb_001500_030 |
0.1227487473 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 7 |
Hb_003747_210 |
0.123246837 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 8 |
Hb_010661_010 |
0.1283254721 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 9 |
Hb_004108_240 |
0.1349956954 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 10 |
Hb_001437_010 |
0.1389331762 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 11 |
Hb_002217_090 |
0.139038797 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 12 |
Hb_000186_200 |
0.1396039522 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 13 |
Hb_000953_050 |
0.1396863887 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 14 |
Hb_000805_230 |
0.1401697283 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 15 |
Hb_003411_030 |
0.1412391757 |
- |
- |
hypothetical protein POPTR_0010s13590g [Populus trichocarpa] |
| 16 |
Hb_138585_100 |
0.1428201529 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 17 |
Hb_005496_180 |
0.1434464322 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 18 |
Hb_006054_030 |
0.1443482831 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 19 |
Hb_002876_250 |
0.145222339 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 20 |
Hb_000252_210 |
0.1456977931 |
- |
- |
PREDICTED: uncharacterized protein LOC105644655 [Jatropha curcas] |