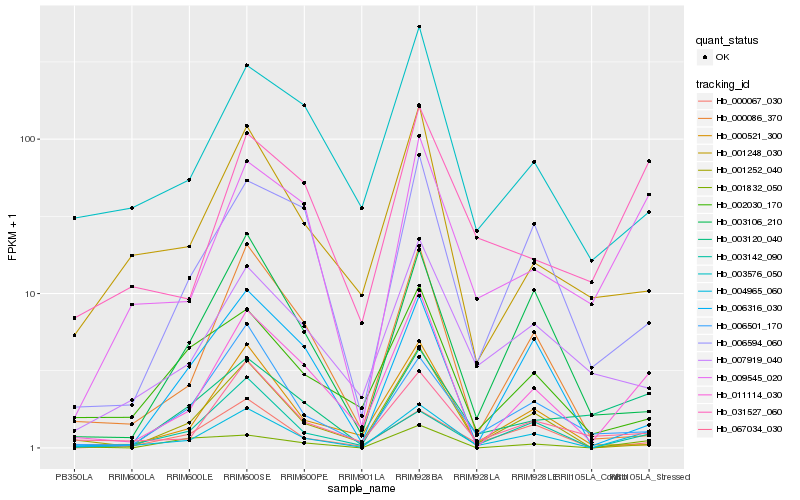
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004965_060 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: U4/U6 small nuclear ribonucleoprotein PRP4-like protein [Jatropha curcas] |
| 2 |
Hb_011114_030 |
0.151459682 |
- |
- |
PREDICTED: probable carboxylesterase 15 [Jatropha curcas] |
| 3 |
Hb_009545_020 |
0.1958547023 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 4 |
Hb_000067_030 |
0.1969276714 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 5 |
Hb_067034_030 |
0.1994019472 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 6 |
Hb_003142_090 |
0.2000358691 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 7 |
Hb_003120_040 |
0.2182552233 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 8 |
Hb_031527_060 |
0.2186164559 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 9 |
Hb_001252_040 |
0.2204780289 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 10 |
Hb_001832_050 |
0.2243761386 |
- |
- |
PREDICTED: cytokinin dehydrogenase 3 [Jatropha curcas] |
| 11 |
Hb_006594_060 |
0.2243792914 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 12 |
Hb_001248_030 |
0.2299529333 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_007919_040 |
0.2300913626 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |
| 14 |
Hb_003106_210 |
0.2304269024 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 15 |
Hb_006501_170 |
0.2311183885 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 16 |
Hb_003576_050 |
0.2357188595 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 17 |
Hb_002030_170 |
0.2363083552 |
- |
- |
PREDICTED: calcium-transporting ATPase 1-like [Jatropha curcas] |
| 18 |
Hb_000521_300 |
0.2367354082 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 19 |
Hb_000086_370 |
0.2367668922 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 20 |
Hb_006316_030 |
0.239441416 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |