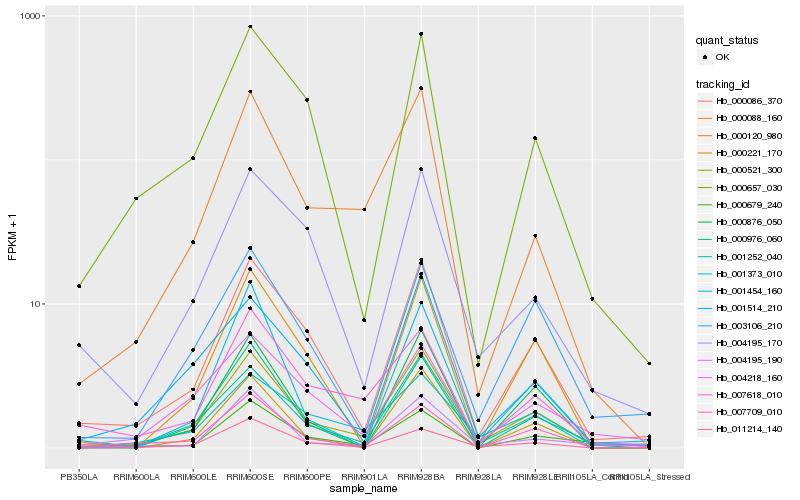
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000521_300 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 2 |
Hb_001252_040 |
0.0982079024 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 3 |
Hb_000086_370 |
0.1084512795 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 4 |
Hb_000088_160 |
0.1333204232 |
- |
- |
PREDICTED: uncharacterized protein LOC105629775 [Jatropha curcas] |
| 5 |
Hb_001373_010 |
0.1461382161 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 6 |
Hb_011214_140 |
0.1479849692 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 7 |
Hb_004195_190 |
0.1484438906 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_007618_010 |
0.1551612326 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 9 |
Hb_007709_010 |
0.1555538105 |
- |
- |
- |
| 10 |
Hb_001514_210 |
0.1563483439 |
- |
- |
hypothetical protein VITISV_012133 [Vitis vinifera] |
| 11 |
Hb_004195_170 |
0.1595324079 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000657_030 |
0.1600074468 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 13 |
Hb_000976_060 |
0.1602657409 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_003106_210 |
0.1607166382 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 15 |
Hb_000876_050 |
0.1609013912 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 16 |
Hb_004218_160 |
0.1609726426 |
- |
- |
NBS-LRR type disease resistance protein [Medicago truncatula] |
| 17 |
Hb_000679_240 |
0.1611696537 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 18 |
Hb_000120_980 |
0.1617344049 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 19 |
Hb_000221_170 |
0.1622629516 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 2 [Jatropha curcas] |
| 20 |
Hb_001454_160 |
0.1629287512 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |