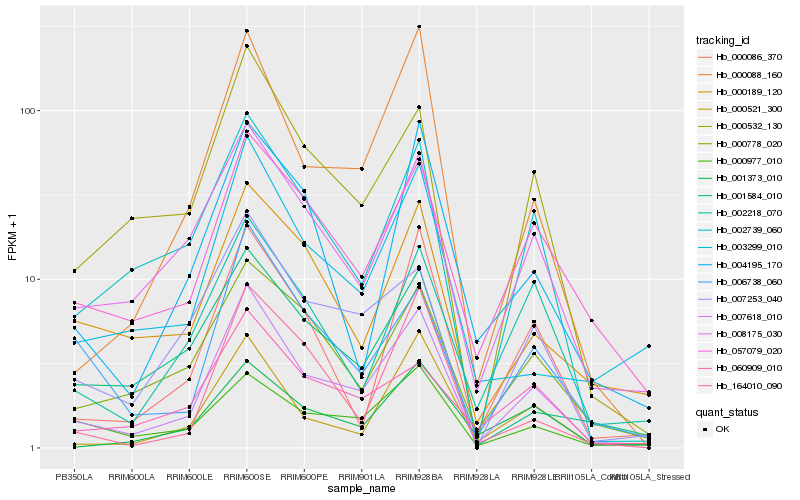
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007618_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 2 |
Hb_008175_030 |
0.1359421913 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 3 |
Hb_000532_130 |
0.144322604 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 4 |
Hb_057079_020 |
0.1447584045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000088_160 |
0.1473573593 |
- |
- |
PREDICTED: uncharacterized protein LOC105629775 [Jatropha curcas] |
| 6 |
Hb_002739_060 |
0.1484787077 |
- |
- |
- |
| 7 |
Hb_007253_040 |
0.1515180858 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000521_300 |
0.1551612326 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 9 |
Hb_000778_020 |
0.1601722847 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 10 |
Hb_003299_010 |
0.1618511857 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 11 |
Hb_000086_370 |
0.1626582825 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 12 |
Hb_000189_120 |
0.1648784828 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 13 |
Hb_000977_010 |
0.1672189916 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 14 |
Hb_001373_010 |
0.1672416772 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_006738_060 |
0.1692747206 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_002218_070 |
0.1698569021 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 17 |
Hb_001584_010 |
0.178037484 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 18 |
Hb_060909_010 |
0.1793366061 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 19 |
Hb_004195_170 |
0.1806758435 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_164010_090 |
0.1814858119 |
- |
- |
cellulose synthase, putative [Ricinus communis] |