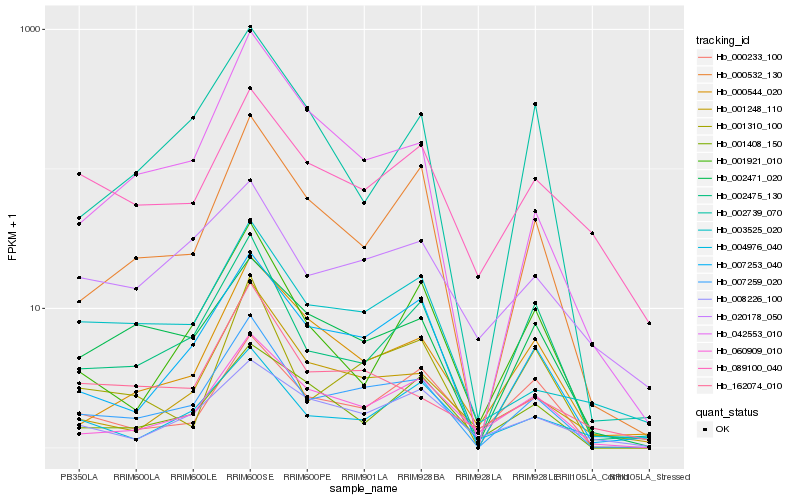
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007259_020 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 2 |
Hb_007253_040 |
0.1260253199 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002475_130 |
0.1352688141 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 4 [Jatropha curcas] |
| 4 |
Hb_000532_130 |
0.14398426 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 5 |
Hb_001310_100 |
0.1547487691 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable N-acetyltransferase HLS1 [Jatropha curcas] |
| 6 |
Hb_060909_010 |
0.1630064695 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 7 |
Hb_001248_110 |
0.1697378848 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 8 |
Hb_002471_020 |
0.1722433336 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 6-like [Jatropha curcas] |
| 9 |
Hb_042553_010 |
0.1758980064 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 10 |
Hb_000544_020 |
0.1764990858 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 11 |
Hb_000233_100 |
0.1799614784 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 12 |
Hb_003525_020 |
0.1817234433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_162074_010 |
0.1864555051 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 14 |
Hb_002739_070 |
0.1875460602 |
- |
- |
hypothetical protein B456_002G169900 [Gossypium raimondii] |
| 15 |
Hb_001921_010 |
0.1882893014 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 16 |
Hb_008226_100 |
0.1930659841 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 17 |
Hb_001408_150 |
0.1937379462 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 18 |
Hb_020178_050 |
0.1963226807 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 19 |
Hb_004976_040 |
0.1967219555 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 20 |
Hb_089100_040 |
0.1978908244 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |