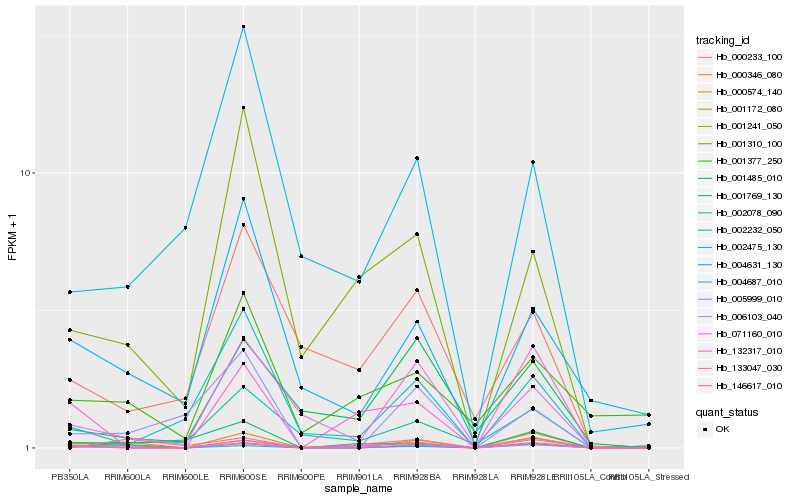
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006103_040 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_001310_100 |
0.2246213471 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable N-acetyltransferase HLS1 [Jatropha curcas] |
| 3 |
Hb_133047_030 |
0.2290694457 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_004687_010 |
0.2474329617 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 5 |
Hb_000574_140 |
0.2628112124 |
- |
- |
- |
| 6 |
Hb_071160_010 |
0.2667420741 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 7 |
Hb_132317_010 |
0.2756597626 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_001241_050 |
0.2781249341 |
- |
- |
PREDICTED: uncharacterized protein LOC104880586 isoform X1 [Vitis vinifera] |
| 9 |
Hb_002232_050 |
0.2858994327 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 10 |
Hb_001485_010 |
0.2890608763 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 11 |
Hb_001377_250 |
0.2951861743 |
- |
- |
dynamin, putative [Ricinus communis] |
| 12 |
Hb_002475_130 |
0.2976643541 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 4 [Jatropha curcas] |
| 13 |
Hb_004631_130 |
0.2984462447 |
- |
- |
helicase domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_000233_100 |
0.2993715322 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 15 |
Hb_001172_080 |
0.2994945391 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 16 |
Hb_000346_080 |
0.3020076996 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 17 |
Hb_146617_010 |
0.302812591 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 18 |
Hb_001769_130 |
0.3057698783 |
- |
- |
- |
| 19 |
Hb_005999_010 |
0.3069655257 |
- |
- |
Sugar transporter ERD6-like 5 [Morus notabilis] |
| 20 |
Hb_002078_090 |
0.3115095808 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |