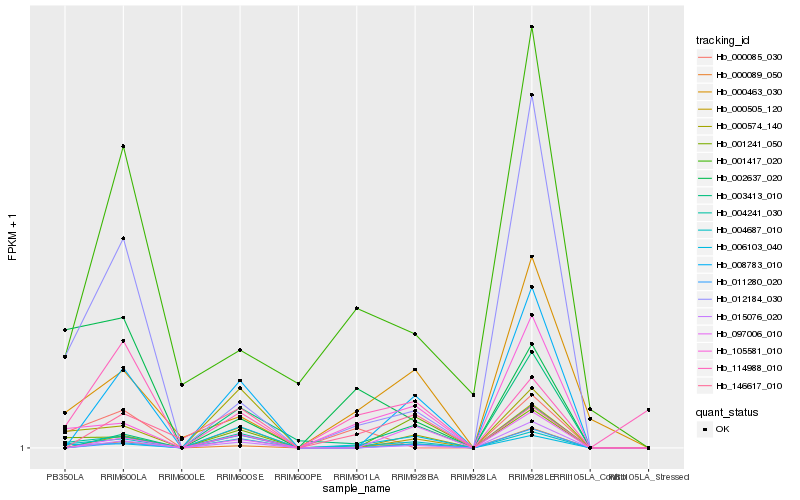
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004687_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 2 |
Hb_000574_140 |
0.2199599272 |
- |
- |
- |
| 3 |
Hb_004241_030 |
0.2293342555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001241_050 |
0.2351854372 |
- |
- |
PREDICTED: uncharacterized protein LOC104880586 isoform X1 [Vitis vinifera] |
| 5 |
Hb_006103_040 |
0.2474329617 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_008783_010 |
0.2665450948 |
- |
- |
- |
| 7 |
Hb_105581_010 |
0.2675735781 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 8 |
Hb_012184_030 |
0.2694996845 |
- |
- |
- |
| 9 |
Hb_000463_030 |
0.2704652897 |
- |
- |
- |
| 10 |
Hb_000505_120 |
0.2844943711 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 11 |
Hb_146617_010 |
0.2910613672 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 12 |
Hb_011280_020 |
0.2967221869 |
- |
- |
polyprotein [Oryza australiensis] |
| 13 |
Hb_000089_050 |
0.3037239678 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_114988_010 |
0.3142568998 |
- |
- |
- |
| 15 |
Hb_003413_010 |
0.316596725 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 16 |
Hb_002637_020 |
0.3231317604 |
- |
- |
- |
| 17 |
Hb_000085_030 |
0.324545974 |
- |
- |
- |
| 18 |
Hb_097006_010 |
0.3255178123 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 19 |
Hb_001417_020 |
0.3283041444 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |
| 20 |
Hb_015076_020 |
0.3286945033 |
- |
- |
IMP dehydrogenase/GMP reductase [Hevea brasiliensis] |