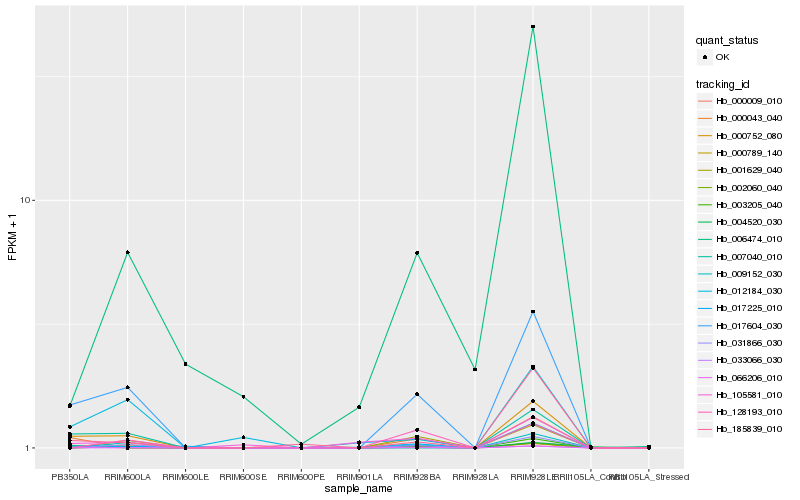
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017604_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000752_080 |
0.1925117939 |
- |
- |
- |
| 3 |
Hb_031866_030 |
0.2416974879 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis melo] |
| 4 |
Hb_002060_040 |
0.2437765849 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 5 |
Hb_009152_030 |
0.2438924824 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 6 |
Hb_033066_030 |
0.2439108731 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 7 |
Hb_000009_010 |
0.2500090721 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 8 |
Hb_012184_030 |
0.2597566265 |
- |
- |
- |
| 9 |
Hb_001629_040 |
0.2604012364 |
- |
- |
hypothetical protein POPTR_0001s41630g [Populus trichocarpa] |
| 10 |
Hb_105581_010 |
0.2846388813 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 11 |
Hb_000789_140 |
0.2872314436 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 12 |
Hb_007040_010 |
0.2873610721 |
- |
- |
Uncharacterized protein TCM_024356 [Theobroma cacao] |
| 13 |
Hb_017225_010 |
0.290141797 |
- |
- |
PREDICTED: nodulation receptor kinase-like [Jatropha curcas] |
| 14 |
Hb_128193_010 |
0.2999862239 |
- |
- |
PREDICTED: uncharacterized protein LOC103699534, partial [Phoenix dactylifera] |
| 15 |
Hb_004520_030 |
0.3024547848 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_185839_010 |
0.30366709 |
- |
- |
- |
| 17 |
Hb_006474_010 |
0.3075548259 |
- |
- |
hypothetical protein VITISV_032357 [Vitis vinifera] |
| 18 |
Hb_000043_040 |
0.308193031 |
- |
- |
CTV.19 [Citrus trifoliata] |
| 19 |
Hb_066206_010 |
0.3086314499 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_003205_040 |
0.3088123983 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |