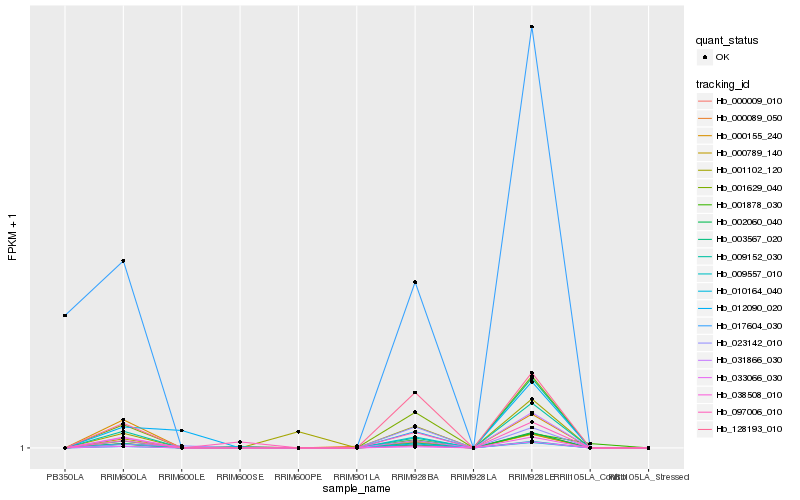
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001629_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s41630g [Populus trichocarpa] |
| 2 |
Hb_000009_010 |
0.0380896308 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 3 |
Hb_128193_010 |
0.082018277 |
- |
- |
PREDICTED: uncharacterized protein LOC103699534, partial [Phoenix dactylifera] |
| 4 |
Hb_031866_030 |
0.1369405002 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis melo] |
| 5 |
Hb_009152_030 |
0.1431435649 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 6 |
Hb_002060_040 |
0.1487168938 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 7 |
Hb_033066_030 |
0.1493643103 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 8 |
Hb_000789_140 |
0.2273163455 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 9 |
Hb_009557_010 |
0.239072037 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 10 |
Hb_097006_010 |
0.2425584184 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 11 |
Hb_010164_040 |
0.2434802077 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_023142_010 |
0.2462249612 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_000155_240 |
0.257880604 |
- |
- |
PREDICTED: uncharacterized protein LOC104888642 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_017604_030 |
0.2604012364 |
- |
- |
- |
| 15 |
Hb_001878_030 |
0.2753431135 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105126050 [Populus euphratica] |
| 16 |
Hb_001102_120 |
0.2881954452 |
- |
- |
- |
| 17 |
Hb_012090_020 |
0.2939319465 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 [Jatropha curcas] |
| 18 |
Hb_038508_010 |
0.2940387316 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 19 |
Hb_003567_020 |
0.2968169519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000089_050 |
0.2991998269 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |