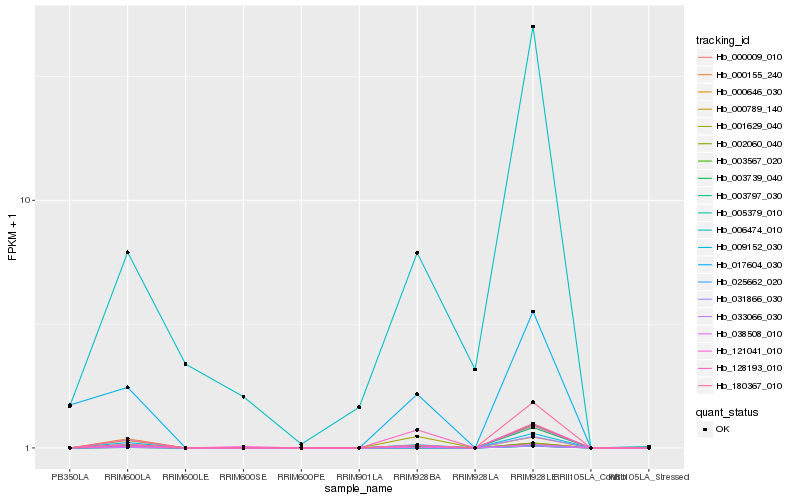
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033066_030 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 2 |
Hb_002060_040 |
0.0007593124 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 3 |
Hb_031866_030 |
0.0146210402 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis melo] |
| 4 |
Hb_009152_030 |
0.0524040154 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 5 |
Hb_000009_010 |
0.1147073665 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 6 |
Hb_001629_040 |
0.1493643103 |
- |
- |
hypothetical protein POPTR_0001s41630g [Populus trichocarpa] |
| 7 |
Hb_000789_140 |
0.169307303 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 8 |
Hb_003567_020 |
0.1799791971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000155_240 |
0.2134438635 |
- |
- |
PREDICTED: uncharacterized protein LOC104888642 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_180367_010 |
0.2192680037 |
- |
- |
PREDICTED: uncharacterized protein LOC105794100 [Gossypium raimondii] |
| 11 |
Hb_006474_010 |
0.2234493804 |
- |
- |
hypothetical protein VITISV_032357 [Vitis vinifera] |
| 12 |
Hb_003739_040 |
0.2241322519 |
- |
- |
RNA-directed DNA polymerase (Reverse transcriptase); Chromo; Zinc finger, CCHC-type; Peptidase aspartic, active site; Polynucleotidyl transferase, Ribonuclease H fold [Medicago truncatula] |
| 13 |
Hb_128193_010 |
0.2292655033 |
- |
- |
PREDICTED: uncharacterized protein LOC103699534, partial [Phoenix dactylifera] |
| 14 |
Hb_121041_010 |
0.2379441461 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |
| 15 |
Hb_017604_030 |
0.2439108731 |
- |
- |
- |
| 16 |
Hb_038508_010 |
0.2509296827 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 17 |
Hb_000646_030 |
0.2589390667 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 18 |
Hb_025662_020 |
0.2589390745 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_003797_030 |
0.2589391486 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_005379_010 |
0.2589394815 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |