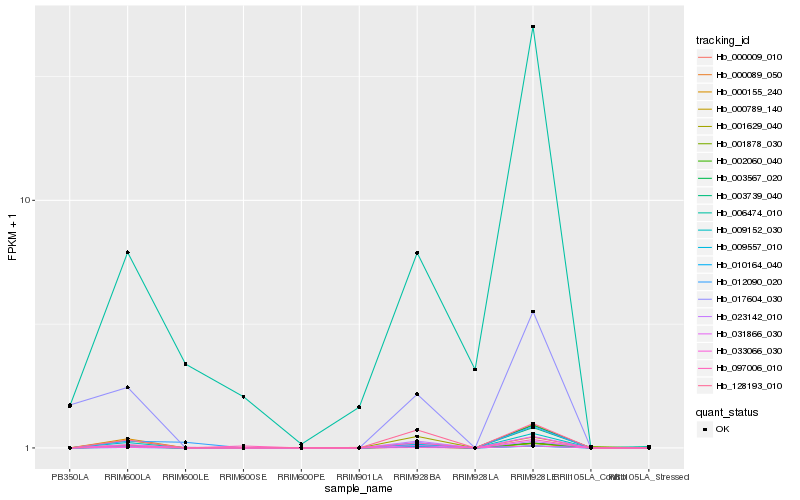
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_010 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 2 |
Hb_001629_040 |
0.0380896308 |
- |
- |
hypothetical protein POPTR_0001s41630g [Populus trichocarpa] |
| 3 |
Hb_031866_030 |
0.1032513521 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis melo] |
| 4 |
Hb_002060_040 |
0.1141029757 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 5 |
Hb_033066_030 |
0.1147073665 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 6 |
Hb_128193_010 |
0.1169570976 |
- |
- |
PREDICTED: uncharacterized protein LOC103699534, partial [Phoenix dactylifera] |
| 7 |
Hb_009152_030 |
0.1171038565 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 8 |
Hb_000789_140 |
0.2163440943 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 9 |
Hb_017604_030 |
0.2500090721 |
- |
- |
- |
| 10 |
Hb_097006_010 |
0.2505740973 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 11 |
Hb_000155_240 |
0.2515166377 |
- |
- |
PREDICTED: uncharacterized protein LOC104888642 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_010164_040 |
0.2570164554 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 13 |
Hb_003567_020 |
0.260707789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_023142_010 |
0.2670800506 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_006474_010 |
0.2731947148 |
- |
- |
hypothetical protein VITISV_032357 [Vitis vinifera] |
| 16 |
Hb_003739_040 |
0.2758668596 |
- |
- |
RNA-directed DNA polymerase (Reverse transcriptase); Chromo; Zinc finger, CCHC-type; Peptidase aspartic, active site; Polynucleotidyl transferase, Ribonuclease H fold [Medicago truncatula] |
| 17 |
Hb_009557_010 |
0.2761596829 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 18 |
Hb_001878_030 |
0.2809228653 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105126050 [Populus euphratica] |
| 19 |
Hb_012090_020 |
0.2832543386 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 [Jatropha curcas] |
| 20 |
Hb_000089_050 |
0.2861462621 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |