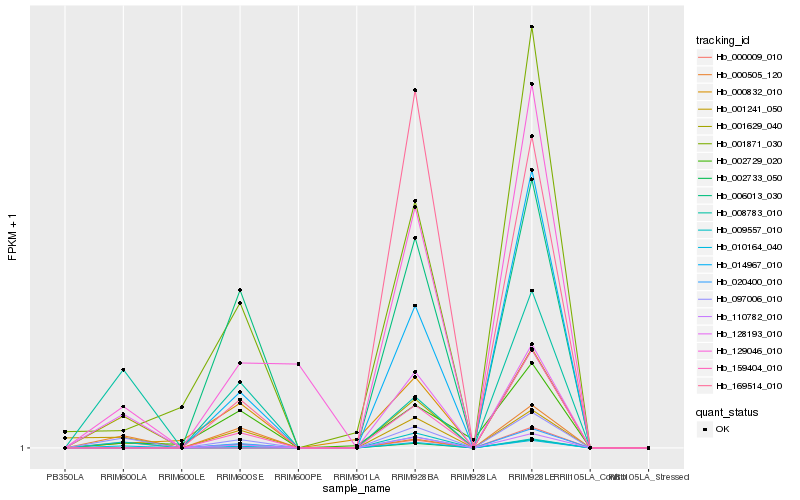
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010164_040 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 2 |
Hb_097006_010 |
0.0473437868 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 3 |
Hb_008783_010 |
0.2231938616 |
- |
- |
- |
| 4 |
Hb_128193_010 |
0.2255054499 |
- |
- |
PREDICTED: uncharacterized protein LOC103699534, partial [Phoenix dactylifera] |
| 5 |
Hb_001241_050 |
0.240755137 |
- |
- |
PREDICTED: uncharacterized protein LOC104880586 isoform X1 [Vitis vinifera] |
| 6 |
Hb_129046_010 |
0.2432730274 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 7 |
Hb_001629_040 |
0.2434802077 |
- |
- |
hypothetical protein POPTR_0001s41630g [Populus trichocarpa] |
| 8 |
Hb_110782_010 |
0.2484517481 |
- |
- |
PREDICTED: uncharacterized protein LOC105797448 [Gossypium raimondii] |
| 9 |
Hb_020400_010 |
0.2538327821 |
- |
- |
PREDICTED: uncharacterized protein LOC105644073 [Jatropha curcas] |
| 10 |
Hb_000009_010 |
0.2570164554 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 11 |
Hb_000505_120 |
0.2649425937 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_000832_010 |
0.2656263548 |
- |
- |
hypothetical protein VITISV_005044 [Vitis vinifera] |
| 13 |
Hb_014967_010 |
0.2675302925 |
- |
- |
- |
| 14 |
Hb_001871_030 |
0.2680986017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_159404_010 |
0.2728470693 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002733_050 |
0.2805468456 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 17 |
Hb_009557_010 |
0.2806639879 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 18 |
Hb_006013_030 |
0.2811839903 |
- |
- |
- |
| 19 |
Hb_169514_010 |
0.2829762384 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 20 |
Hb_002729_020 |
0.2835003104 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |