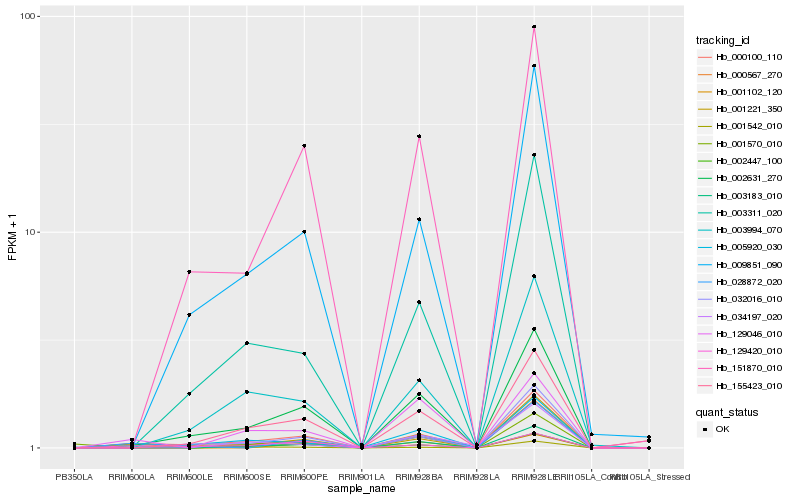
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003183_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105785048, partial [Gossypium raimondii] |
| 2 |
Hb_129046_010 |
0.1482837179 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 3 |
Hb_001221_350 |
0.1532024664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_028872_020 |
0.1867697425 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 5 |
Hb_002631_270 |
0.1920515574 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s09930g [Populus trichocarpa] |
| 6 |
Hb_002447_100 |
0.1930821272 |
- |
- |
hypothetical protein POPTR_0009s06260g [Populus trichocarpa] |
| 7 |
Hb_155423_010 |
0.2013118803 |
- |
- |
- |
| 8 |
Hb_000567_270 |
0.2027807394 |
- |
- |
hypothetical protein L484_017162 [Morus notabilis] |
| 9 |
Hb_003311_020 |
0.2249631501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001102_120 |
0.2256060575 |
- |
- |
- |
| 11 |
Hb_000100_110 |
0.2276758638 |
- |
- |
- |
| 12 |
Hb_003994_070 |
0.2301707961 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 13 |
Hb_009851_090 |
0.2302399919 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 14 |
Hb_001542_010 |
0.2323185722 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 15 |
Hb_032016_010 |
0.2351149796 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040-like isoform X1 [Citrus sinensis] |
| 16 |
Hb_151870_010 |
0.235474694 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_034197_020 |
0.2378644175 |
- |
- |
hypothetical protein VITISV_041099 [Vitis vinifera] |
| 18 |
Hb_129420_010 |
0.240535474 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 19 |
Hb_005920_030 |
0.2411656403 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 20 |
Hb_001570_010 |
0.2419229685 |
- |
- |
- |