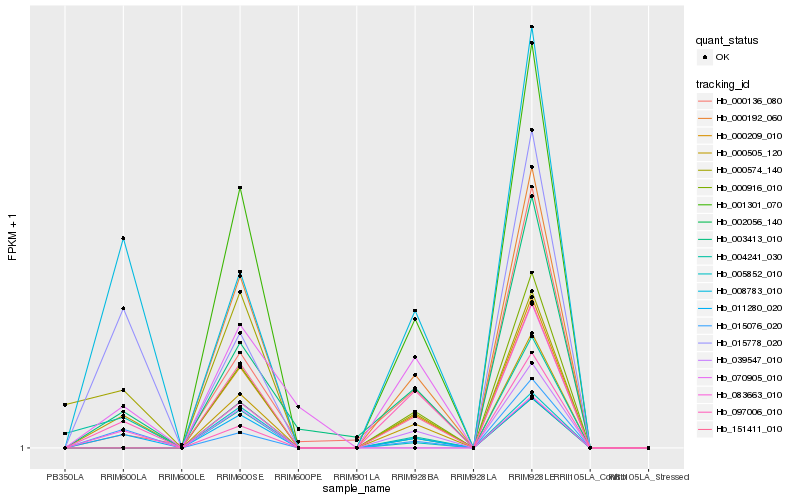
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011280_020 |
0.0 |
- |
- |
polyprotein [Oryza australiensis] |
| 2 |
Hb_000505_120 |
0.0709279503 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_008783_010 |
0.1591431908 |
- |
- |
- |
| 4 |
Hb_015076_020 |
0.1942311773 |
- |
- |
IMP dehydrogenase/GMP reductase [Hevea brasiliensis] |
| 5 |
Hb_003413_010 |
0.2097877233 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 6 |
Hb_083663_010 |
0.2223941873 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 7 |
Hb_004241_030 |
0.2301108104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000574_140 |
0.2330994047 |
- |
- |
- |
| 9 |
Hb_000136_080 |
0.2410889427 |
- |
- |
hypothetical protein AALP_AA8G499800 [Arabis alpina] |
| 10 |
Hb_151411_010 |
0.2587851575 |
- |
- |
- |
| 11 |
Hb_000192_060 |
0.2601684335 |
- |
- |
- |
| 12 |
Hb_000209_010 |
0.2613151243 |
- |
- |
- |
| 13 |
Hb_001301_070 |
0.2637325888 |
- |
- |
- |
| 14 |
Hb_002056_140 |
0.2653070906 |
- |
- |
- |
| 15 |
Hb_015778_020 |
0.2659215159 |
- |
- |
Putative methyltransferase family protein [Solanum demissum] |
| 16 |
Hb_000916_010 |
0.2667659132 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 17 |
Hb_039547_010 |
0.2669188791 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 18 |
Hb_005852_010 |
0.2698195746 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 19 |
Hb_070905_010 |
0.2713900611 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 20 |
Hb_097006_010 |
0.2736884262 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |