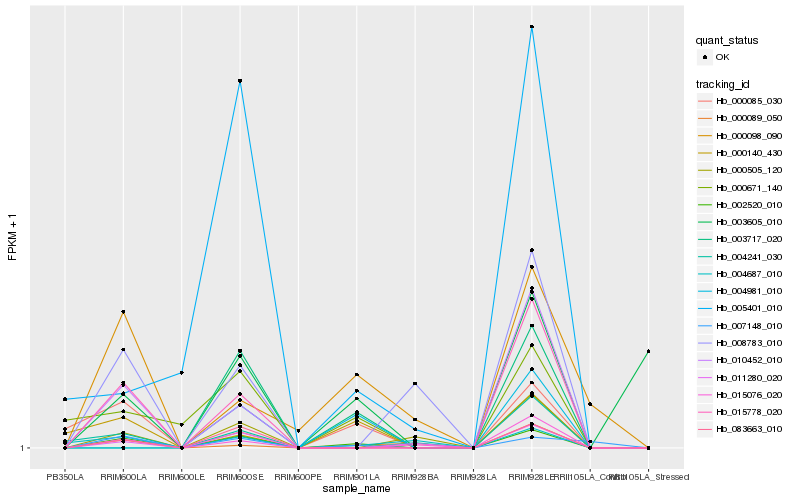
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002520_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 2 |
Hb_083663_010 |
0.2489487746 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 3 |
Hb_015778_020 |
0.2497477859 |
- |
- |
Putative methyltransferase family protein [Solanum demissum] |
| 4 |
Hb_010452_010 |
0.2750215315 |
- |
- |
PREDICTED: uncharacterized protein LOC104232369 [Nicotiana sylvestris] |
| 5 |
Hb_000085_030 |
0.3054366822 |
- |
- |
- |
| 6 |
Hb_000671_140 |
0.3082250222 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 7 |
Hb_000140_430 |
0.3109932517 |
- |
- |
- |
| 8 |
Hb_007148_010 |
0.3114047816 |
- |
- |
PREDICTED: uncharacterized protein LOC103427080 [Malus domestica] |
| 9 |
Hb_011280_020 |
0.3228945685 |
- |
- |
polyprotein [Oryza australiensis] |
| 10 |
Hb_000505_120 |
0.3323419237 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 11 |
Hb_015076_020 |
0.3357172216 |
- |
- |
IMP dehydrogenase/GMP reductase [Hevea brasiliensis] |
| 12 |
Hb_005401_010 |
0.3389065701 |
- |
- |
- |
| 13 |
Hb_000089_050 |
0.3417152197 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_000098_090 |
0.3467278974 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 15 |
Hb_003717_020 |
0.3468472221 |
- |
- |
- |
| 16 |
Hb_004687_010 |
0.3498300061 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 17 |
Hb_003605_010 |
0.3516859775 |
- |
- |
- |
| 18 |
Hb_008783_010 |
0.3525883418 |
- |
- |
- |
| 19 |
Hb_004241_030 |
0.3560612555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004981_010 |
0.359807544 |
- |
- |
- |