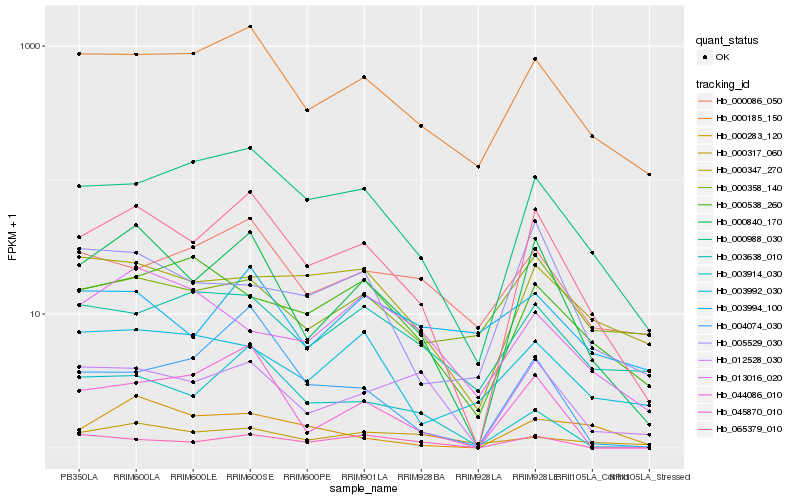
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000840_170 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 2 |
Hb_065379_010 |
0.0945746323 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 3 |
Hb_000185_150 |
0.1940290992 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 4 |
Hb_012528_030 |
0.1980256101 |
- |
- |
- |
| 5 |
Hb_044086_010 |
0.2012271156 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000988_030 |
0.2130681784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005529_030 |
0.2332923731 |
- |
- |
PREDICTED: cytochrome P450 94A1-like [Populus euphratica] |
| 8 |
Hb_045870_010 |
0.2386725077 |
- |
- |
- |
| 9 |
Hb_003914_030 |
0.2414943502 |
- |
- |
- |
| 10 |
Hb_000317_060 |
0.2484838306 |
- |
- |
PREDICTED: uncharacterized protein LOC105647051 [Jatropha curcas] |
| 11 |
Hb_003992_030 |
0.2494704626 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic [Gossypium arboreum] |
| 12 |
Hb_000283_120 |
0.2519928313 |
- |
- |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 13 |
Hb_003638_010 |
0.2525454929 |
transcription factor |
TF Family: ARID |
PREDICTED: lysine-specific demethylase 5B isoform X2 [Jatropha curcas] |
| 14 |
Hb_004074_030 |
0.2538449779 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000347_270 |
0.2548256787 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 16 |
Hb_000358_140 |
0.2550187911 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_013016_020 |
0.255699276 |
- |
- |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 18 |
Hb_000538_260 |
0.255779812 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 19 |
Hb_003994_100 |
0.2571326496 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000086_050 |
0.2574298286 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 1 [Jatropha curcas] |