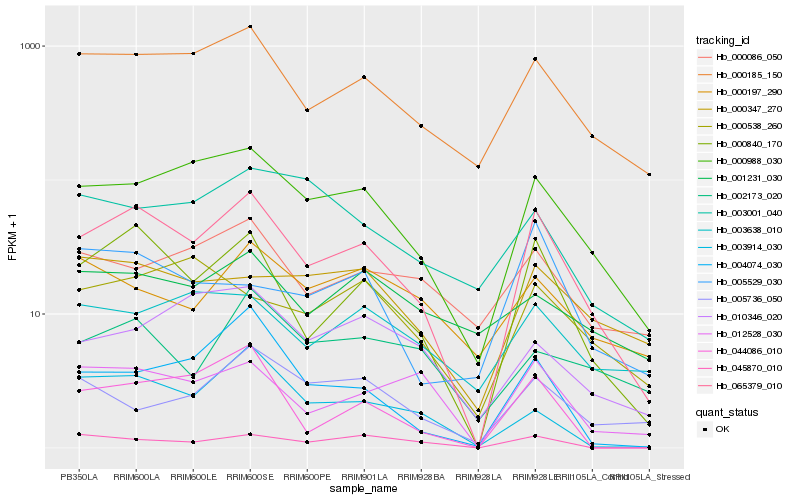
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065379_010 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 2 |
Hb_000840_170 |
0.0945746323 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 3 |
Hb_000988_030 |
0.1409961437 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000185_150 |
0.1539712556 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 5 |
Hb_012528_030 |
0.1932661986 |
- |
- |
- |
| 6 |
Hb_044086_010 |
0.1964135561 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_004074_030 |
0.2016875138 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_003914_030 |
0.2084710834 |
- |
- |
- |
| 9 |
Hb_000347_270 |
0.2112264999 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 10 |
Hb_000086_050 |
0.2132217037 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 1 [Jatropha curcas] |
| 11 |
Hb_045870_010 |
0.2156071452 |
- |
- |
- |
| 12 |
Hb_003638_010 |
0.216102359 |
transcription factor |
TF Family: ARID |
PREDICTED: lysine-specific demethylase 5B isoform X2 [Jatropha curcas] |
| 13 |
Hb_000538_260 |
0.2216855103 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 14 |
Hb_002173_020 |
0.2240741839 |
- |
- |
PREDICTED: uncharacterized protein LOC105801078 [Gossypium raimondii] |
| 15 |
Hb_010346_020 |
0.2242229362 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 16 |
Hb_003001_040 |
0.2247085687 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 17 |
Hb_001231_030 |
0.2255395046 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 18 |
Hb_005736_050 |
0.2256320937 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 19 |
Hb_000197_290 |
0.2267882961 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 20 |
Hb_005529_030 |
0.2277243762 |
- |
- |
PREDICTED: cytochrome P450 94A1-like [Populus euphratica] |