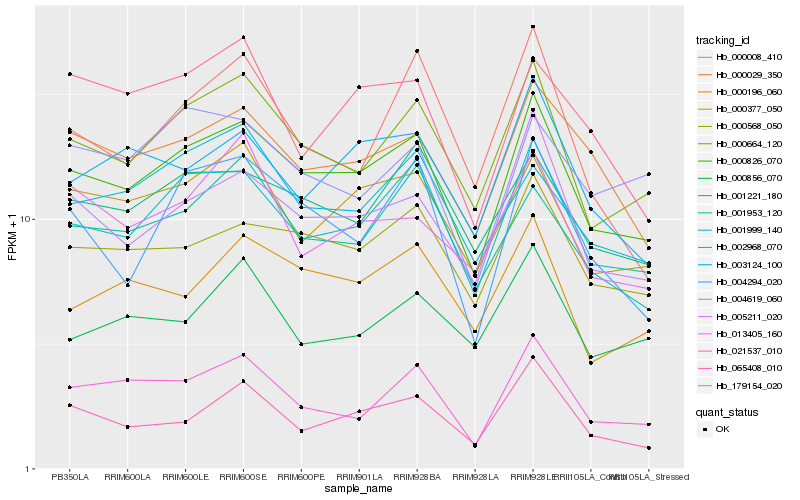
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021537_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000826_070 |
0.0920388096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000664_120 |
0.095950754 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001999_140 |
0.0987147979 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_001953_120 |
0.118164318 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005211_020 |
0.119220095 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000029_350 |
0.1193888476 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 8 |
Hb_000856_070 |
0.1210555683 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 9 |
Hb_000377_050 |
0.1215270835 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 10 |
Hb_065408_010 |
0.1220464063 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000008_410 |
0.1226467986 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 12 |
Hb_000568_050 |
0.123554709 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004619_060 |
0.125938314 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 14 |
Hb_003124_100 |
0.1266271292 |
- |
- |
- |
| 15 |
Hb_002968_070 |
0.1282677221 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 16 |
Hb_001221_180 |
0.1285914049 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 17 |
Hb_013405_160 |
0.1317118507 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_179154_020 |
0.131745463 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 19 |
Hb_004294_020 |
0.1317718051 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000196_060 |
0.1327326574 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |