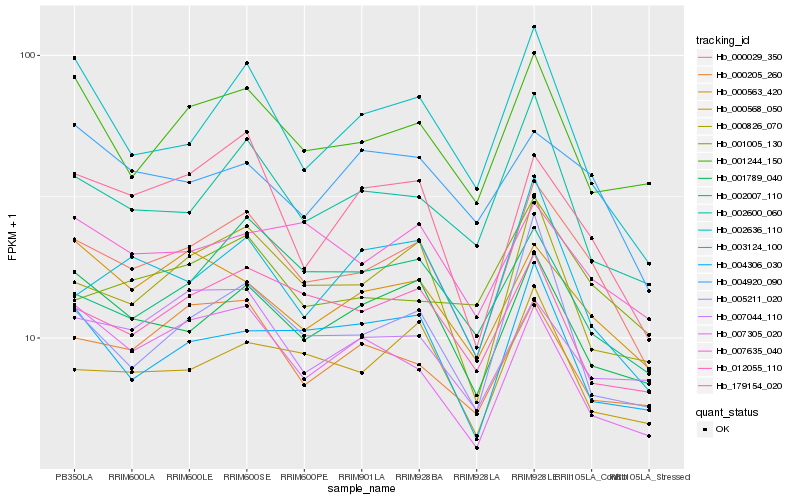
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000029_350 |
0.0 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 2 |
Hb_000826_070 |
0.0851187914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001789_040 |
0.086117436 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 4 |
Hb_179154_020 |
0.0895961327 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 5 |
Hb_002600_060 |
0.0922772835 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 6 |
Hb_000568_050 |
0.0923150154 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001244_150 |
0.0952462108 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 8 |
Hb_000563_420 |
0.0953368444 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 9 |
Hb_012055_110 |
0.0967554727 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 10 |
Hb_007635_040 |
0.0970262097 |
- |
- |
PREDICTED: WAT1-related protein At3g45870 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005211_020 |
0.0977442958 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003124_100 |
0.0979165735 |
- |
- |
- |
| 13 |
Hb_002007_110 |
0.0980927327 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001005_130 |
0.0993148582 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 15 |
Hb_002636_110 |
0.1005130558 |
- |
- |
PREDICTED: heat shock protein 83-like isoform X2 [Gossypium raimondii] |
| 16 |
Hb_000205_260 |
0.1005233857 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 17 |
Hb_004920_090 |
0.1009836456 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_007044_110 |
0.1027874399 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_004306_030 |
0.1030699911 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 20 |
Hb_007305_020 |
0.1032793914 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |