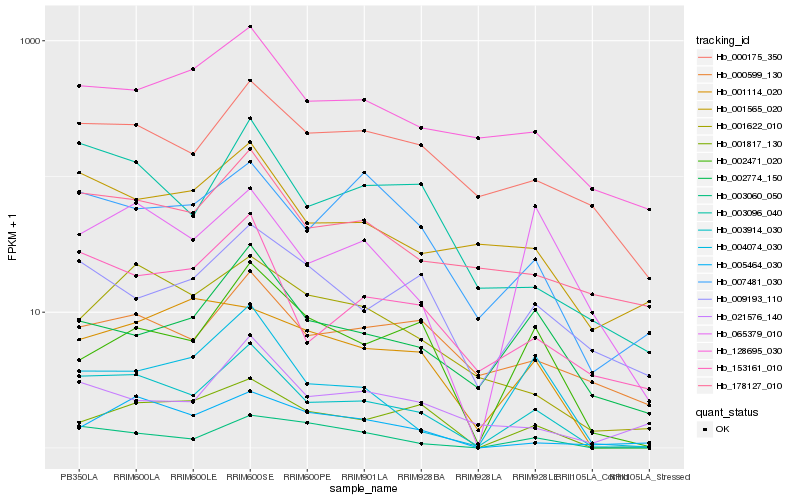
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003914_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_003096_040 |
0.1702072654 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0005s07290g [Populus trichocarpa] |
| 3 |
Hb_153161_010 |
0.1711075733 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 4 |
Hb_003060_050 |
0.1876472924 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 5 |
Hb_001817_130 |
0.1900600208 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 isoform X3 [Cucumis melo] |
| 6 |
Hb_178127_010 |
0.1911806605 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 7 |
Hb_001565_020 |
0.193961903 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial, partial [Jatropha curcas] |
| 8 |
Hb_004074_030 |
0.1942255341 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_007481_030 |
0.1977111248 |
- |
- |
PREDICTED: uncharacterized protein LOC105628433 [Jatropha curcas] |
| 10 |
Hb_000175_350 |
0.2032769337 |
- |
- |
phopholipase d alpha, putative [Ricinus communis] |
| 11 |
Hb_001622_010 |
0.2034035826 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_128695_030 |
0.2046753591 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 13 |
Hb_002471_020 |
0.2064615615 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 6-like [Jatropha curcas] |
| 14 |
Hb_065379_010 |
0.2084710834 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 15 |
Hb_002774_150 |
0.2088862469 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 16 |
Hb_001114_020 |
0.2098315421 |
desease resistance |
Gene Name: LRR_4 |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 17 |
Hb_021576_140 |
0.2160095211 |
- |
- |
PREDICTED: protein POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION-like [Jatropha curcas] |
| 18 |
Hb_005464_030 |
0.2160936928 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_009193_110 |
0.2168787176 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000599_130 |
0.2182185543 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |