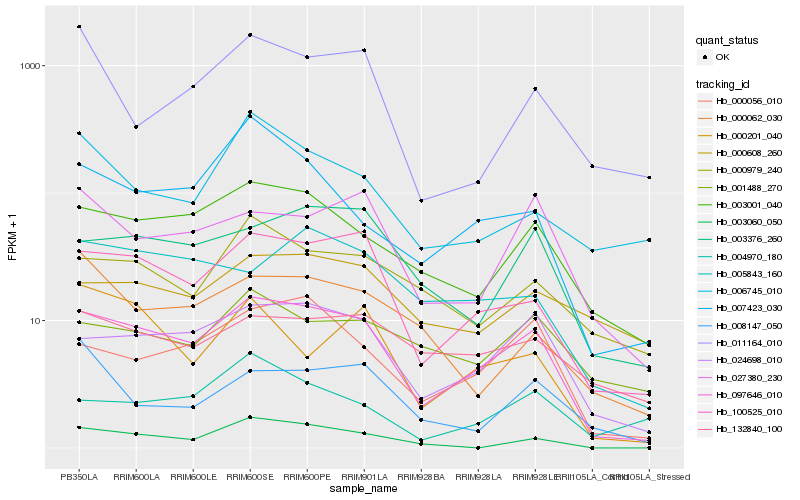
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_097646_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 2 |
Hb_100525_010 |
0.1176646192 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 3 |
Hb_024698_010 |
0.1178502156 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46 [Jatropha curcas] |
| 4 |
Hb_003001_040 |
0.1416738208 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 5 |
Hb_000056_010 |
0.1497609032 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 6 |
Hb_003376_260 |
0.1613879981 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 7 |
Hb_027380_230 |
0.1729272166 |
- |
- |
PREDICTED: CSC1-like protein ERD4 [Jatropha curcas] |
| 8 |
Hb_000979_240 |
0.1835570367 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 9 |
Hb_132840_100 |
0.184238563 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 10 |
Hb_005843_160 |
0.185766822 |
- |
- |
PREDICTED: uncharacterized protein LOC105112024 [Populus euphratica] |
| 11 |
Hb_007423_030 |
0.1862998993 |
- |
- |
PREDICTED: protein EXORDIUM-like 2 [Jatropha curcas] |
| 12 |
Hb_003060_050 |
0.1872566498 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 13 |
Hb_001488_270 |
0.1937436884 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 14 |
Hb_011164_010 |
0.19606203 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 15 |
Hb_000062_030 |
0.1990768995 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000608_260 |
0.2000465713 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 17 |
Hb_000201_040 |
0.2042477992 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 18 |
Hb_004970_180 |
0.2046151511 |
- |
- |
- |
| 19 |
Hb_006745_010 |
0.2051299686 |
- |
- |
PREDICTED: uncharacterized protein LOC105640095 [Jatropha curcas] |
| 20 |
Hb_008147_050 |
0.2054744899 |
- |
- |
PREDICTED: uncharacterized protein LOC105646845 [Jatropha curcas] |