| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
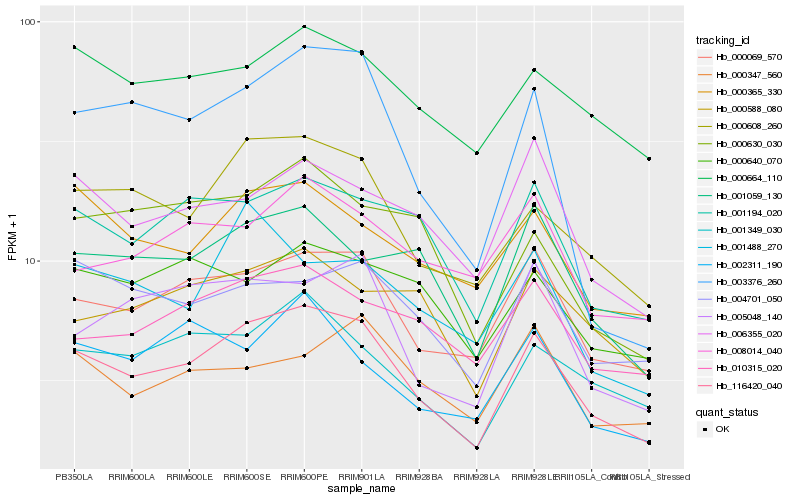
Hb_116420_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000608_260 |
0.0903663786 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 3 |
Hb_000069_570 |
0.091582816 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001349_030 |
0.109206443 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001194_020 |
0.1092837677 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 6 |
Hb_005048_140 |
0.1170905811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001059_130 |
0.1188086421 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000365_330 |
0.1190344542 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_003376_260 |
0.1190846587 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000664_110 |
0.1206052843 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 11 |
Hb_010315_020 |
0.1237000958 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 12 |
Hb_004701_050 |
0.12396503 |
- |
- |
alpha1 family protein [Populus trichocarpa] |
| 13 |
Hb_006355_020 |
0.1241884679 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 14 |
Hb_000630_030 |
0.1244212622 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 15 |
Hb_000588_080 |
0.1266142359 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 16 |
Hb_002311_190 |
0.1303627058 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 17 |
Hb_001488_270 |
0.1314707808 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 18 |
Hb_000640_070 |
0.132233696 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 19 |
Hb_008014_040 |
0.1327675883 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 20 |
Hb_000347_560 |
0.13541721 |
- |
- |
- |