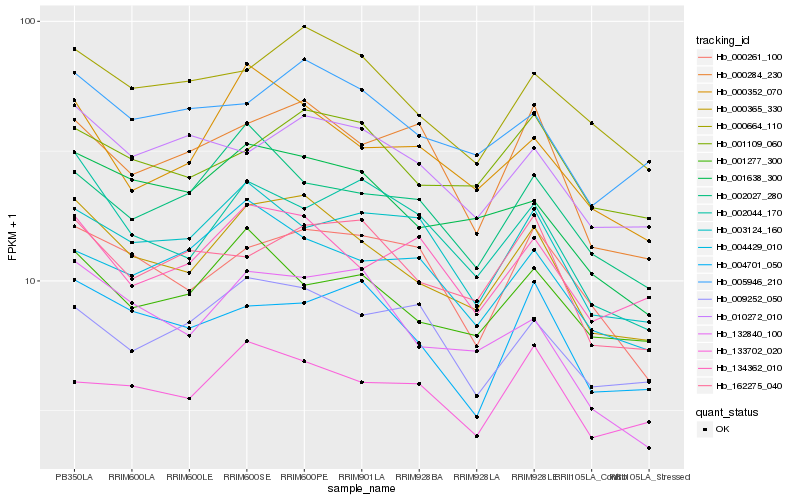
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_330 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_001638_300 |
0.0783341147 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 3 |
Hb_000664_110 |
0.0823152984 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 4 |
Hb_005946_210 |
0.0864897762 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 5 |
Hb_000352_070 |
0.0893423238 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 6 |
Hb_132840_100 |
0.0923856964 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 7 |
Hb_001109_060 |
0.0935239182 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_134362_010 |
0.0938411355 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 9 |
Hb_002044_170 |
0.0947593463 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 10 |
Hb_001277_300 |
0.094872334 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 11 |
Hb_010272_010 |
0.0951052495 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 12 |
Hb_000284_230 |
0.0956154723 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 13 |
Hb_162275_040 |
0.0962154202 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004701_050 |
0.1000372625 |
- |
- |
alpha1 family protein [Populus trichocarpa] |
| 15 |
Hb_002027_280 |
0.1003948905 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 16 |
Hb_004429_010 |
0.1004494061 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 17 |
Hb_009252_050 |
0.1015622399 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_000261_100 |
0.1015776359 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 19 |
Hb_133702_020 |
0.1020637018 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_003124_160 |
0.1020775486 |
- |
- |
dynamin, putative [Ricinus communis] |