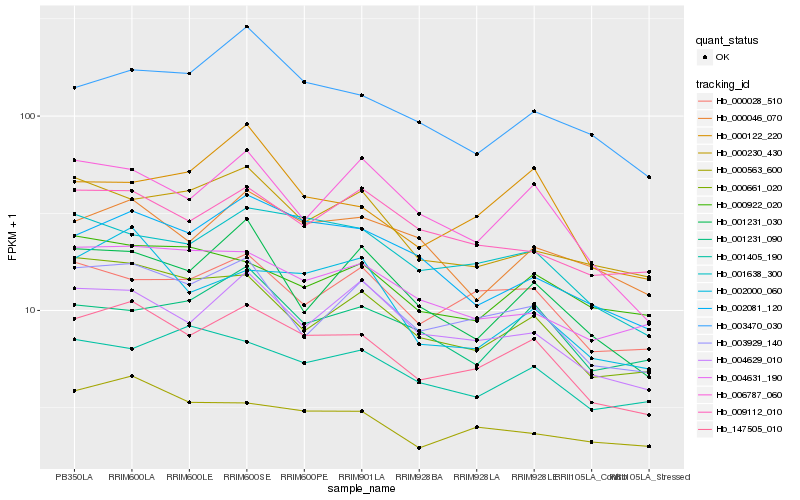
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_147505_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 2 |
Hb_003929_140 |
0.080287564 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_001638_300 |
0.0829782277 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 4 |
Hb_004629_010 |
0.0893933762 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 5 |
Hb_000661_020 |
0.0898159532 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 6 |
Hb_000563_600 |
0.0902325759 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 7 |
Hb_000028_510 |
0.0937517256 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 8 |
Hb_002000_060 |
0.094353272 |
- |
- |
PREDICTED: protein LIKE COV 2 [Jatropha curcas] |
| 9 |
Hb_002081_120 |
0.094431477 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 10 |
Hb_004631_190 |
0.097701748 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 11 |
Hb_006787_060 |
0.0981242004 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000046_070 |
0.1010505488 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 13 |
Hb_009112_010 |
0.1019349566 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 14 |
Hb_003470_030 |
0.1023639461 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 15 |
Hb_000230_430 |
0.1058213656 |
- |
- |
catalytic, putative [Ricinus communis] |
| 16 |
Hb_001231_030 |
0.1063331438 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 17 |
Hb_001405_190 |
0.1064750289 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000122_220 |
0.1074881264 |
- |
- |
Prenylated Rab acceptor protein, putative [Ricinus communis] |
| 19 |
Hb_001231_090 |
0.1077521202 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000922_020 |
0.1082565103 |
- |
- |
protein binding protein, putative [Ricinus communis] |