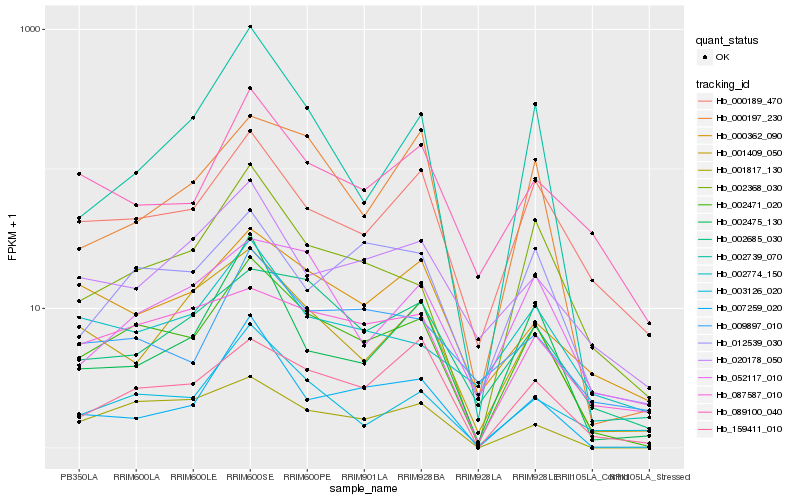
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002471_020 |
0.0 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 6-like [Jatropha curcas] |
| 2 |
Hb_000189_470 |
0.149896696 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |
| 3 |
Hb_001817_130 |
0.1500608525 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 isoform X3 [Cucumis melo] |
| 4 |
Hb_002368_030 |
0.1556493252 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 5 |
Hb_003126_020 |
0.1627902354 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 6 |
Hb_159411_010 |
0.1711148642 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 7 |
Hb_001409_050 |
0.172096091 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_007259_020 |
0.1722433336 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 9 |
Hb_012539_030 |
0.1731636668 |
- |
- |
PREDICTED: uncharacterized protein LOC105638247 [Jatropha curcas] |
| 10 |
Hb_089100_040 |
0.1740636673 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 11 |
Hb_002685_030 |
0.175019216 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |
| 12 |
Hb_009897_010 |
0.175745444 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 13 |
Hb_087587_010 |
0.1762201321 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 14 |
Hb_002774_150 |
0.1793499845 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 15 |
Hb_000362_090 |
0.1795526001 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000197_230 |
0.1824285201 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 17 |
Hb_052117_010 |
0.1856384593 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_020178_050 |
0.1858363591 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 19 |
Hb_002739_070 |
0.1870917806 |
- |
- |
hypothetical protein B456_002G169900 [Gossypium raimondii] |
| 20 |
Hb_002475_130 |
0.1871242425 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 4 [Jatropha curcas] |