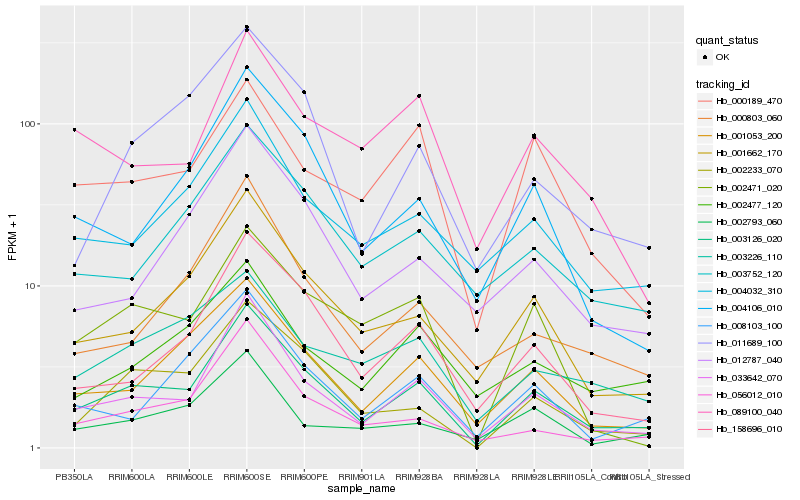
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003126_020 |
0.0 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 2 |
Hb_033642_070 |
0.0898954648 |
- |
- |
PREDICTED: oligopeptide transporter 7-like [Jatropha curcas] |
| 3 |
Hb_001053_200 |
0.1356246086 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 4 |
Hb_001662_170 |
0.1358672641 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 5 |
Hb_002233_070 |
0.1372983255 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_158696_010 |
0.1424064145 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 7 |
Hb_004106_010 |
0.1509697285 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004032_310 |
0.1518090593 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 9 |
Hb_056012_010 |
0.1549985322 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 10 |
Hb_003752_120 |
0.15713704 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008103_100 |
0.1578210576 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 12 |
Hb_011689_100 |
0.1578729836 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 13 |
Hb_002471_020 |
0.1627902354 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 6-like [Jatropha curcas] |
| 14 |
Hb_000803_060 |
0.1638880509 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 15 |
Hb_012787_040 |
0.1640226317 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002477_120 |
0.1673763142 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 17 |
Hb_003226_110 |
0.1679161207 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 18 |
Hb_000189_470 |
0.1695962638 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |
| 19 |
Hb_089100_040 |
0.1720978725 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 20 |
Hb_002793_060 |
0.1721115789 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g02980 [Populus euphratica] |