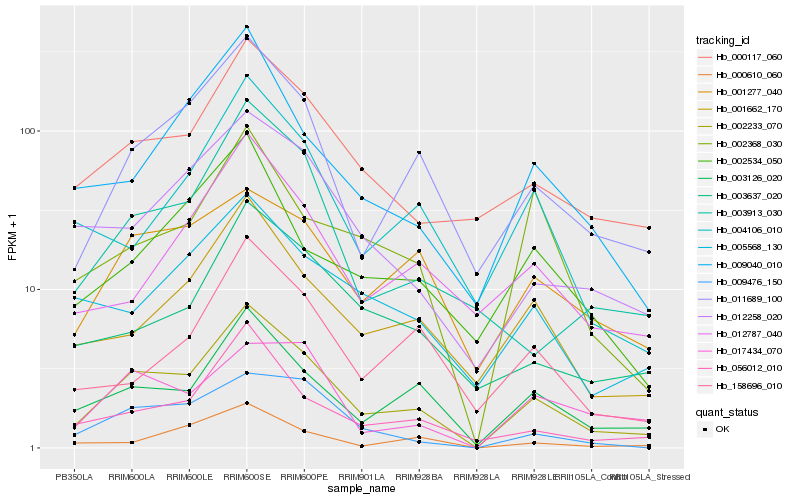
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002233_070 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_003126_020 |
0.1372983255 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 3 |
Hb_011689_100 |
0.1419830804 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 4 |
Hb_000117_060 |
0.1596432178 |
- |
- |
PREDICTED: uncharacterized protein LOC103949199 [Pyrus x bretschneideri] |
| 5 |
Hb_056012_010 |
0.1647729253 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 6 |
Hb_001662_170 |
0.1664161103 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 7 |
Hb_009476_150 |
0.1763420199 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 8 |
Hb_012258_020 |
0.1775281644 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 9 |
Hb_009040_010 |
0.1810814368 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_001277_040 |
0.1827788795 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 11 |
Hb_003637_020 |
0.1831254638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004106_010 |
0.1832843401 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003913_030 |
0.1833959577 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 14 |
Hb_012787_040 |
0.1862448903 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 15 |
Hb_005568_130 |
0.1876666192 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_002534_050 |
0.1883328247 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 17 |
Hb_002368_030 |
0.188676634 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 18 |
Hb_158696_010 |
0.1906428743 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 19 |
Hb_017434_070 |
0.192135218 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_000610_060 |
0.1931621259 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |