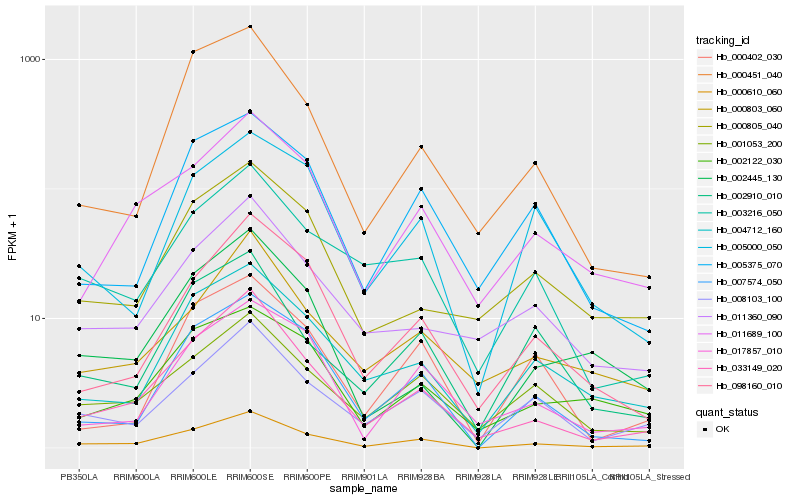
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000610_060 |
0.0 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 2 |
Hb_033149_020 |
0.1027322137 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 3 |
Hb_007574_050 |
0.1274367131 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 4 |
Hb_008103_100 |
0.1283568792 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 5 |
Hb_004712_160 |
0.1320085205 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 6 |
Hb_011689_100 |
0.139433407 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 7 |
Hb_001053_200 |
0.1403503349 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 8 |
Hb_098160_010 |
0.1483153303 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_002122_030 |
0.1507317919 |
- |
- |
- |
| 10 |
Hb_017857_010 |
0.1530125006 |
- |
- |
- |
| 11 |
Hb_002445_130 |
0.1530343826 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 12 |
Hb_003216_050 |
0.154116174 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 13 |
Hb_002910_010 |
0.1546456259 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 14 |
Hb_005375_070 |
0.1557076169 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 15 |
Hb_000451_040 |
0.156937517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000803_060 |
0.161301634 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 17 |
Hb_005000_050 |
0.1628428535 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 18 |
Hb_000402_030 |
0.1643226184 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 19 |
Hb_000805_040 |
0.1652711524 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 20 |
Hb_011360_090 |
0.1657317563 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |