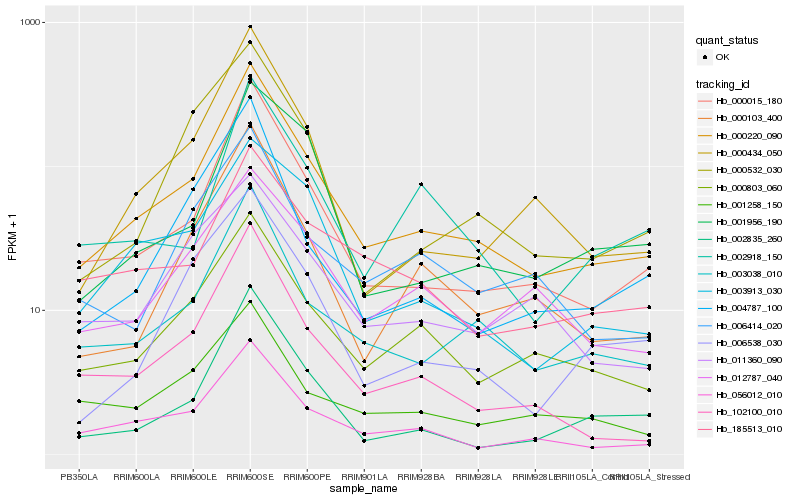
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000220_090 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 2 |
Hb_000015_180 |
0.0889394987 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 3 |
Hb_003038_010 |
0.1037068136 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_056012_010 |
0.1150645739 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 5 |
Hb_000803_060 |
0.118452898 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 6 |
Hb_102100_010 |
0.1301596904 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 7 |
Hb_000434_050 |
0.1321631674 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 8 |
Hb_000103_400 |
0.1338563425 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003913_030 |
0.1390041465 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 10 |
Hb_185513_010 |
0.144219481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_012787_040 |
0.1459940583 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002835_260 |
0.1473496778 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 13 |
Hb_000532_030 |
0.1485684477 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |
| 14 |
Hb_006414_020 |
0.1489302966 |
transcription factor |
TF Family: MYB |
- |
| 15 |
Hb_001956_190 |
0.1504622497 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 16 |
Hb_001258_150 |
0.1535344381 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 17 |
Hb_006538_030 |
0.1536374477 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 18 |
Hb_004787_100 |
0.1550791936 |
- |
- |
PREDICTED: transcription factor bHLH47 [Jatropha curcas] |
| 19 |
Hb_002918_150 |
0.1561600581 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 20 |
Hb_011360_090 |
0.1606061993 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |