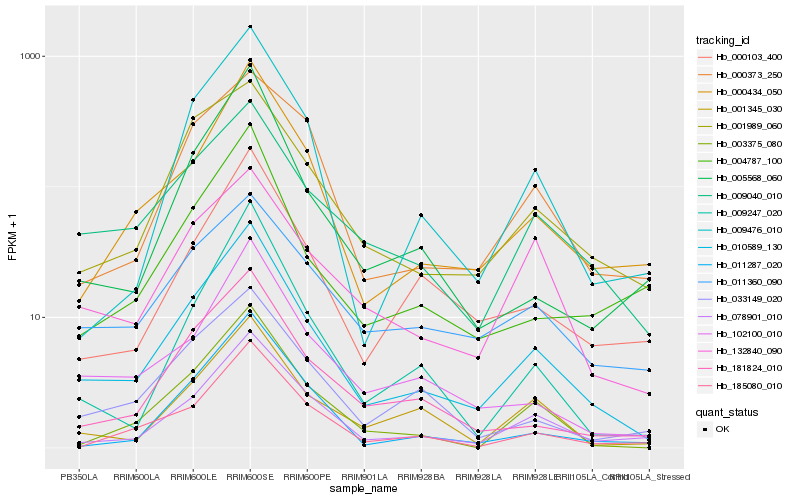
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010589_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_078901_010 |
0.1201483968 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 3 |
Hb_000434_050 |
0.1282597715 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 4 |
Hb_009040_010 |
0.1328950806 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_003375_080 |
0.1371629294 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 6 |
Hb_102100_010 |
0.1389726035 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 7 |
Hb_181824_010 |
0.1402787384 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_001989_060 |
0.1420365241 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001345_030 |
0.1447205249 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 10 |
Hb_009476_010 |
0.1461790521 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 11 |
Hb_185080_010 |
0.1484867985 |
- |
- |
- |
| 12 |
Hb_000103_400 |
0.1509817793 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_009247_020 |
0.1533882859 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 14 |
Hb_033149_020 |
0.1554954302 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 15 |
Hb_132840_090 |
0.1564219801 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005568_060 |
0.1569843363 |
- |
- |
PREDICTED: mitogen-activated protein kinase 3 [Jatropha curcas] |
| 17 |
Hb_011360_090 |
0.157840165 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 18 |
Hb_011287_020 |
0.1603046723 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_004787_100 |
0.1609293544 |
- |
- |
PREDICTED: transcription factor bHLH47 [Jatropha curcas] |
| 20 |
Hb_000373_250 |
0.1620846004 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |