| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
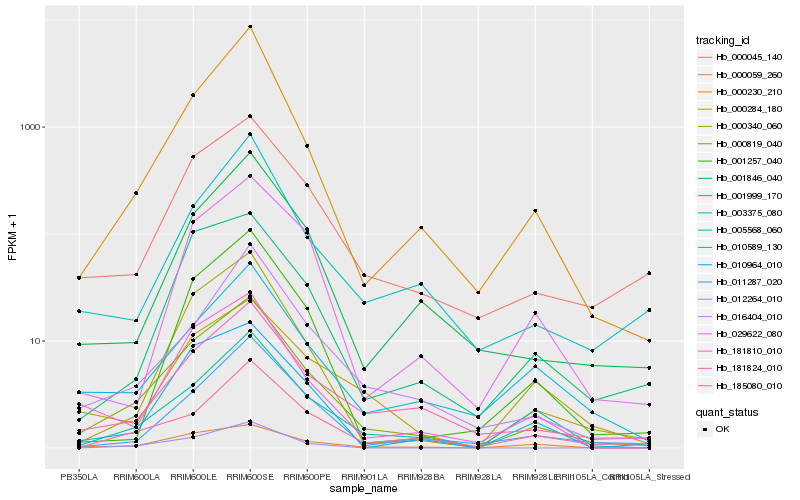
Hb_000819_040 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_000045_140 |
0.132252736 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 3 |
Hb_000230_210 |
0.1370463324 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 4 |
Hb_181824_010 |
0.1404303452 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 5 |
Hb_000340_060 |
0.1406925878 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_181810_010 |
0.1412104889 |
- |
- |
hypothetical protein JCGZ_14167 [Jatropha curcas] |
| 7 |
Hb_003375_080 |
0.1461504098 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 8 |
Hb_010964_010 |
0.1465247706 |
- |
- |
- |
| 9 |
Hb_001846_040 |
0.1476386518 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_000059_260 |
0.1484470252 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: peroxidase 60 [Jatropha curcas] |
| 11 |
Hb_016404_010 |
0.1486474023 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 12 |
Hb_000284_180 |
0.1539661583 |
- |
- |
hypothetical protein EUGRSUZ_B01865 [Eucalyptus grandis] |
| 13 |
Hb_011287_020 |
0.1540899068 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_012264_010 |
0.1556752774 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001999_170 |
0.160532156 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 16 |
Hb_185080_010 |
0.1624877633 |
- |
- |
- |
| 17 |
Hb_029622_080 |
0.1649042643 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 18 |
Hb_005568_060 |
0.1672307572 |
- |
- |
PREDICTED: mitogen-activated protein kinase 3 [Jatropha curcas] |
| 19 |
Hb_001257_040 |
0.1677683251 |
- |
- |
PREDICTED: uncharacterized protein LOC105640055 [Jatropha curcas] |
| 20 |
Hb_010589_130 |
0.1681137868 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |