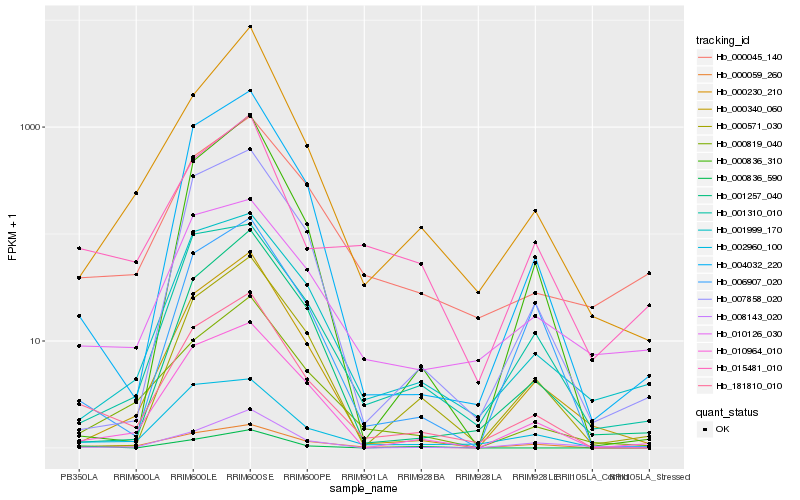
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_181810_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_14167 [Jatropha curcas] |
| 2 |
Hb_010964_010 |
0.137798833 |
- |
- |
- |
| 3 |
Hb_000819_040 |
0.1412104889 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_004032_220 |
0.1417659999 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 5 |
Hb_000340_060 |
0.1494723943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000059_260 |
0.150252715 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: peroxidase 60 [Jatropha curcas] |
| 7 |
Hb_002960_100 |
0.1550128745 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 8 |
Hb_007858_020 |
0.1608151689 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 9 |
Hb_001999_170 |
0.1619392975 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 10 |
Hb_000045_140 |
0.1638190105 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 11 |
Hb_015481_010 |
0.1644137714 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 12 |
Hb_000230_210 |
0.1654193092 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 13 |
Hb_008143_020 |
0.1660171478 |
- |
- |
PREDICTED: rhomboid-like protease 2 [Jatropha curcas] |
| 14 |
Hb_006907_020 |
0.1669878224 |
- |
- |
1-amino-cyclopropane-1-carboxylic acid oxidase 3 [Manihot esculenta] |
| 15 |
Hb_001310_010 |
0.1680063446 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |
| 16 |
Hb_000836_310 |
0.1700677462 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 30 [Jatropha curcas] |
| 17 |
Hb_001257_040 |
0.1706350216 |
- |
- |
PREDICTED: uncharacterized protein LOC105640055 [Jatropha curcas] |
| 18 |
Hb_000571_030 |
0.1712455521 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 19 |
Hb_000836_590 |
0.1752470596 |
- |
- |
hypothetical protein JCGZ_14318 [Jatropha curcas] |
| 20 |
Hb_010126_030 |
0.1756429086 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |