| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
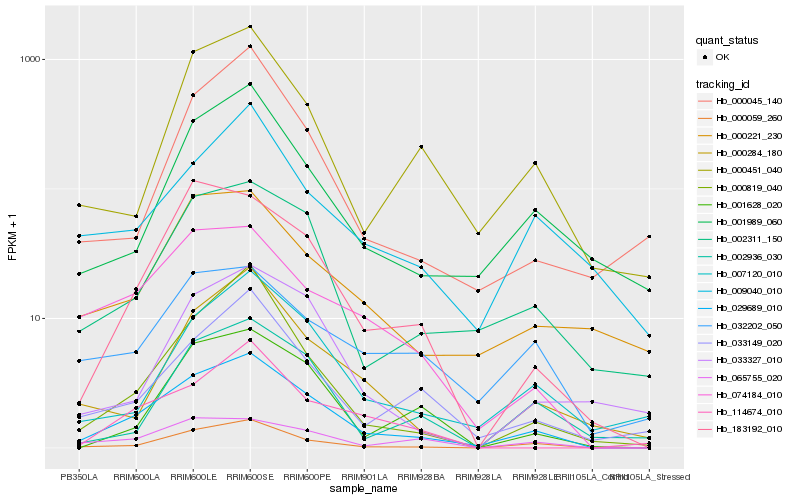
Hb_029689_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 2 |
Hb_000059_260 |
0.132679465 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: peroxidase 60 [Jatropha curcas] |
| 3 |
Hb_002311_150 |
0.1622850489 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 4 |
Hb_183192_010 |
0.1732747908 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |
| 5 |
Hb_074184_010 |
0.1760964409 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 6 |
Hb_007120_010 |
0.1765295827 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |
| 7 |
Hb_033149_020 |
0.1783106516 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 8 |
Hb_000819_040 |
0.178877381 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_065755_020 |
0.1808512203 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 10 |
Hb_001989_060 |
0.1841935159 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000451_040 |
0.1870482835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001628_020 |
0.1872965977 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 13 |
Hb_000284_180 |
0.1901513084 |
- |
- |
hypothetical protein EUGRSUZ_B01865 [Eucalyptus grandis] |
| 14 |
Hb_002936_030 |
0.1982501314 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000221_230 |
0.1985540658 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 16 |
Hb_114674_010 |
0.2015024649 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 17 |
Hb_000045_140 |
0.2019135465 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 18 |
Hb_032202_050 |
0.2022218621 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_033327_010 |
0.2027013327 |
- |
- |
SYM8 [Pisum sativum] |
| 20 |
Hb_009040_010 |
0.2030802253 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |