| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
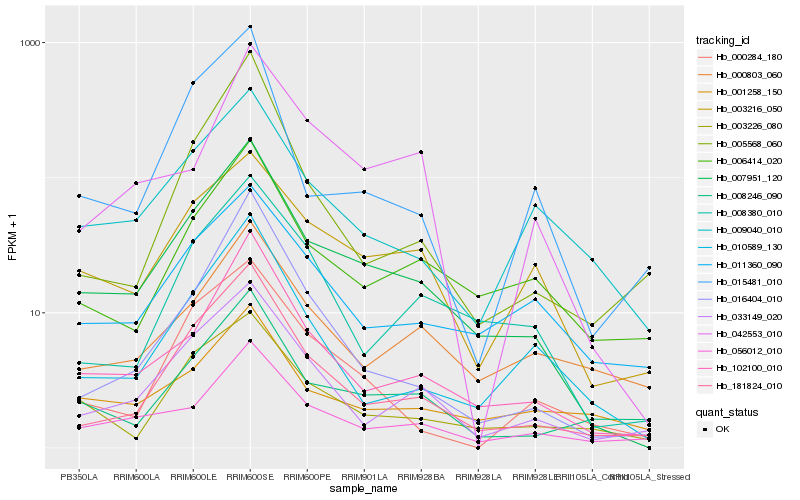
Hb_007951_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 2 |
Hb_102100_010 |
0.1267708252 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_181824_010 |
0.1337219219 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 4 |
Hb_033149_020 |
0.1527277453 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 5 |
Hb_056012_010 |
0.1572070696 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 6 |
Hb_006414_020 |
0.1632581154 |
transcription factor |
TF Family: MYB |
- |
| 7 |
Hb_003216_050 |
0.1633029729 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 8 |
Hb_008246_090 |
0.1636978331 |
- |
- |
- |
| 9 |
Hb_003226_080 |
0.1665600889 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5-like [Jatropha curcas] |
| 10 |
Hb_008380_010 |
0.1667009619 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 11 |
Hb_016404_010 |
0.1776380938 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 12 |
Hb_010589_130 |
0.1781070598 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_009040_010 |
0.1781364314 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_011360_090 |
0.1804294719 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 15 |
Hb_001258_150 |
0.185617997 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 16 |
Hb_015481_010 |
0.1878098262 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 17 |
Hb_042553_010 |
0.1884648542 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 18 |
Hb_000284_180 |
0.1888916704 |
- |
- |
hypothetical protein EUGRSUZ_B01865 [Eucalyptus grandis] |
| 19 |
Hb_000803_060 |
0.1897478561 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 20 |
Hb_005568_060 |
0.1924771122 |
- |
- |
PREDICTED: mitogen-activated protein kinase 3 [Jatropha curcas] |