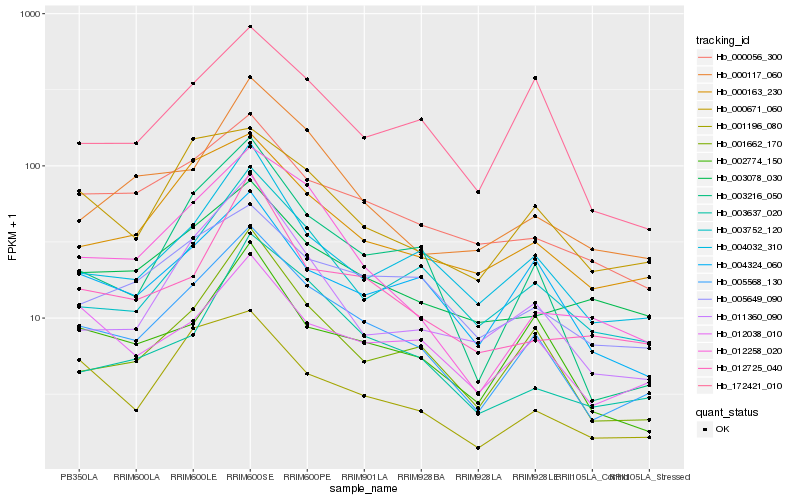
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005568_130 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_003216_050 |
0.1081388789 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 3 |
Hb_001662_170 |
0.1128609997 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 4 |
Hb_000163_230 |
0.124832474 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 5 |
Hb_002774_150 |
0.126463766 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 6 |
Hb_004032_310 |
0.1269748195 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 7 |
Hb_003752_120 |
0.1280056821 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000056_300 |
0.1314344608 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_012258_020 |
0.1317830415 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 10 |
Hb_004324_060 |
0.1323549093 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 11 |
Hb_005649_090 |
0.1368282841 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 12 |
Hb_011360_090 |
0.1392026829 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 13 |
Hb_012038_010 |
0.1401562752 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 14 |
Hb_000117_060 |
0.1402337909 |
- |
- |
PREDICTED: uncharacterized protein LOC103949199 [Pyrus x bretschneideri] |
| 15 |
Hb_172421_010 |
0.1403741295 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 16 |
Hb_001196_080 |
0.1425698354 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 17 |
Hb_003078_030 |
0.1434469502 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 18 |
Hb_003637_020 |
0.1439513978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000671_060 |
0.1454047132 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_012725_040 |
0.1455133176 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |